Website management
| 1. Constructing/Removing a website (by using a command line interface) | |
|---|---|
| (1) Preparing configuration files | |
| (1-1) With assemblies | |
| (1-1-1) Preparing a configuraion file | |
# Website name (Required)
>Website_name Example # Input assemblies (Required) ### Path list for files in FASTA format
> Assemblies Human ../data/example_inputs/human.fa Mouse ../data/example_inputs/mouse.fa Cow ../data/example_inputs/cow.fa # Divergence times (Required) ### Example) Human-chimpanzee: near, Human-mouse: medium, Human-chicken: far
> Divtimes Human,Mouse medium Human,Cow medium Mouse,Cow medium # Resolutions (Required) ### The minimum size of synteny blocks in base pair
> Resolutions
150000,300000,400000,500000 # Gene annotations of reference (Optional) ### Path list of files in Gene transfer format(GTF)
>Annotation Human ../data/example_inputs/Homo_sapiens.GRCh38.87.gtf.gz Mouse ../data/example_inputs/Mus_musculus.GRCm38.87.gtf.gz #Cytogenetic bands (Optional)
> Cytoband Human ../data/example_inputs/Human.cytoband Mouse ../data/example_inputs/mouse.cytoband Cow ../data/example_inputs/mouse.cytoband # Pre-built circos plots (Optional) ### If user wants to visualize all chromosomes/scaffolds, user should write 'all' instead of specific chromosomes.
> Circos1 resolution:150000 Human:chr1,chr3,chr5,chr6,chr8,chr12,chr15,chr19 Mouse:chr1,chr3,chr7,chr17,chr18 Cow:chr3,chr9,chr10,chr14,chr17,chr22 # Email address (Optional) ### It is used to make a contact link in published website.
> Email
User@email.com | |
| (1-1-2) Required input data & formats | |
>1 TTATTCCGCATCTTCTGAAGAAGATGTTCCGAATATATCCTTAGAAAGGA GGTGATCCAGCCGCACCTTCCGATACGGCTACCTTGTTACGACTTCACCC
1 havana gene 11869 14409 . + . gene_id "ENSG00000223972"; gene_version "5"; gene_name "DDX11L1"; gene_source "havana"; gene_biotype "transcribed_unprocessed_pseudogene";
havana_gene "OTTHUMG00000000961"; havana_gene_version "2"; 1 havana transcript 11869 14409 . + . gene_id "ENSG00000223972"; gene_version "5"; transcript_id "ENST00000456328"; transcript_version "2"; gene_name "DDX11L1"; gene_source "havana"; gene_biotype "transcribed_unprocessed_pseudogene"; havana_gene "OTTHUMG00000000961"; havana_gene_version "2"; transcript_name "DDX11L1-002"; transcript_source "havana"; transcript_biotype "processed_transcript"; havana_transcript "OTTHUMT00000362751"; havana_transcript_version "1"; tag "basic"; transcript_support_level "1";
chr1 0 2300000 p36.33 gneg chr1 2300000 5400000 p36.32 gpos25 chr1 5400000 7200000 p36.31 gneg | |
| (1-2) With synteny blocks | |
| (1-2-1) Preparing a configuraion file | |
# Website name (Required)
> Website_name Example # Input synteny blocks (Required) ### Path list of files in synteny format
> Synteny_blocks Human,Mouse ../data/example_inputs/human.mouse.synteny Human,Cow ../data/example_inputs/human.cow.synteny # Genome size files (Required) ### Path list of files in genome size format
> Genome_size Human ../data/example_inputs/human.sizes Mouse ../data/example_inputs/mouse.sizes Cow ../data/example_inputs/cow.sizes # Gene annotations of reference (Optional) ### Path list of files in Gene transfer format(GTF)
> Annotation Human ../data/example_inputs/Homo_sapiens.GRCh38.87.gtf.gz Mouse ../data/example_inputs/Mus_musculus.GRCm38.87.gtf.gz # Cytogenetic bands (Optional)
> Cytoband Human ../data/example_inputs/Human.cytoband Mouse ../data/example_inputs/mouse.cytoband Cow ../data/example_inputs/mouse.cytoband # Pre-built circos plots (Optional) ### If user wants to visualize all chromosomes/scaffolds, user should write 'all' instead of specific chromosomes.
> Circos1 resolution:150000 Human:chr1,chr3,chr5,chr6,chr8,chr12,chr15,chr19 Mouse:chr1,chr3,chr7,chr17,chr18 Cow:chr3,chr9,chr10,chr14,chr17,chr22 # Email address (Optional) ###It is used to make a contact link in published website.
> Email
User@email.com | |
| (1-2-2) Required input data & formats | |
# Synteny format One line starting with a '>' sign, followd by the synteny numberThe other two lines containing the coordinates and the orientation in each scaffold of pairwise alignment (First line is only for reference chromosome(scaffold)
>1 Human.chr1:933237-58547094 + Mouse.chr4:103313481-156255944 - >2 Human.chr1:58654678-67136459 + Mouse.chr4:94941999-103299247 + # Genome size format First column containing chromosome(scaffold) name and second column containing chromosome(scaffold) sizeHow to make? [mySyntenyPortal_root]/src/third_party/kent/faSize -detailed [FASTA] > [size file]
chr1 249250621 chr2 243199373
1 havana gene 11869 14409 . + . gene_id "ENSG00000223972"; gene_version "5"; gene_name "DDX11L1"; gene_source "havana"; gene_biotype "transcribed_unprocessed_pseudogene";
havana_gene "OTTHUMG00000000961"; havana_gene_version "2"; 1 havana transcript 11869 14409 . + . gene_id "ENSG00000223972"; gene_version "5"; transcript_id "ENST00000456328"; transcript_version "2"; gene_name "DDX11L1"; gene_source "havana"; gene_biotype "transcribed_unprocessed_pseudogene"; havana_gene "OTTHUMG00000000961"; havana_gene_version "2"; transcript_name "DDX11L1-002"; transcript_source "havana"; transcript_biotype "processed_transcript"; havana_transcript "OTTHUMT00000362751"; havana_transcript_version "1"; tag "basic"; transcript_support_level "1";
chr1 0 2300000 p36.33 gneg chr1 2300000 5400000 p36.32 gpos25 chr1 5400000 7200000 p36.31 gneg | |
| (2) Command lines | |
| (2-1) Building website | |
User can use the 'mySyntenyPortal' perl script to build website. Command: ./mySyntenyPortal build <parameters> Parameter: -conf|c=> Configuration file -core|p=> Number of threads (default: 10) Example: ./mySyntenyPortal build -p 10 -conf ./configurations/sample.conf
| |
| (2-2) Removing website | |
User can use the 'mySyntenyPortal' perl script to remove website. Command: ./mySyntenyPortal remove <parameters> Parameter: -website_name|w=> Website name -conf|c=> Configuration file Example: ./mySyntenyPortal remove -website_name Sample_website
| |
| 2. Publishing/Unpublishing a website (by using a web interface) | |
|---|---|
| (1) Drawing default plots | |
|
This step is to draw plots which are used as default plots in the pubilshed website. 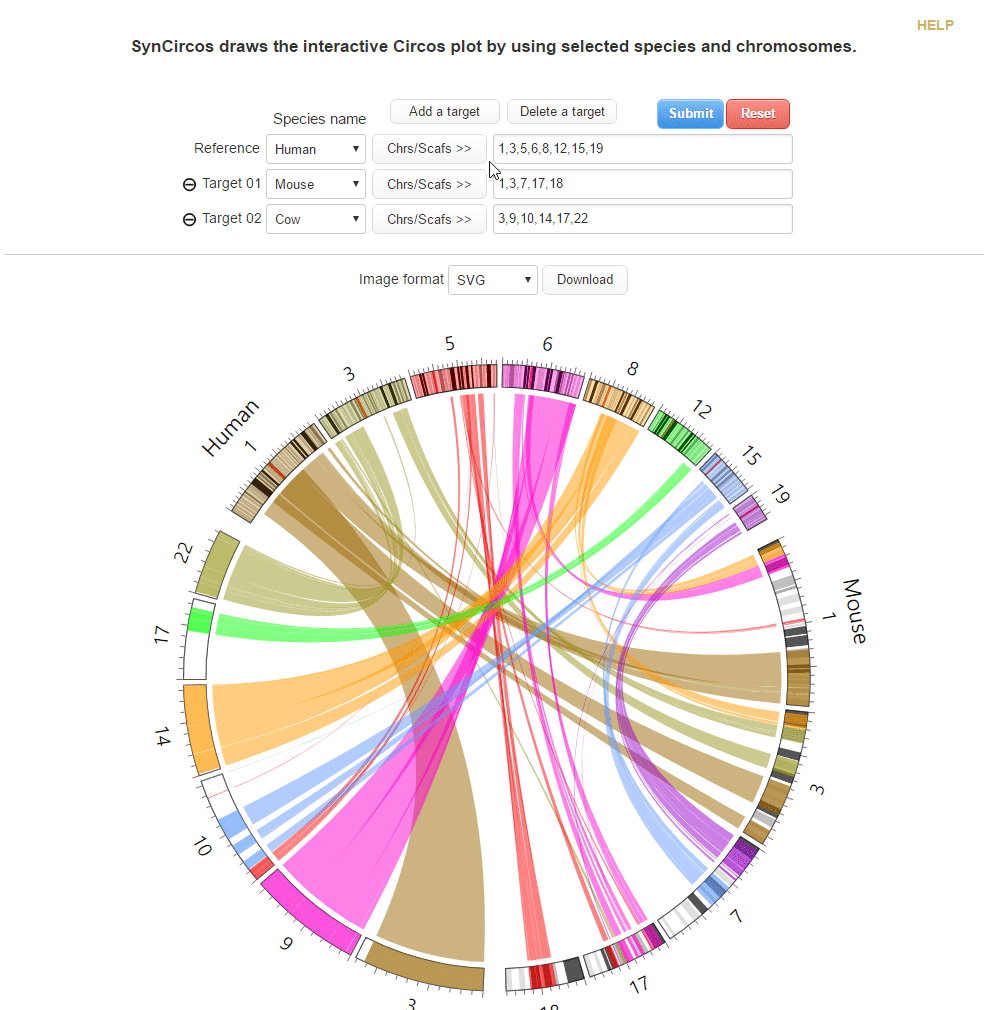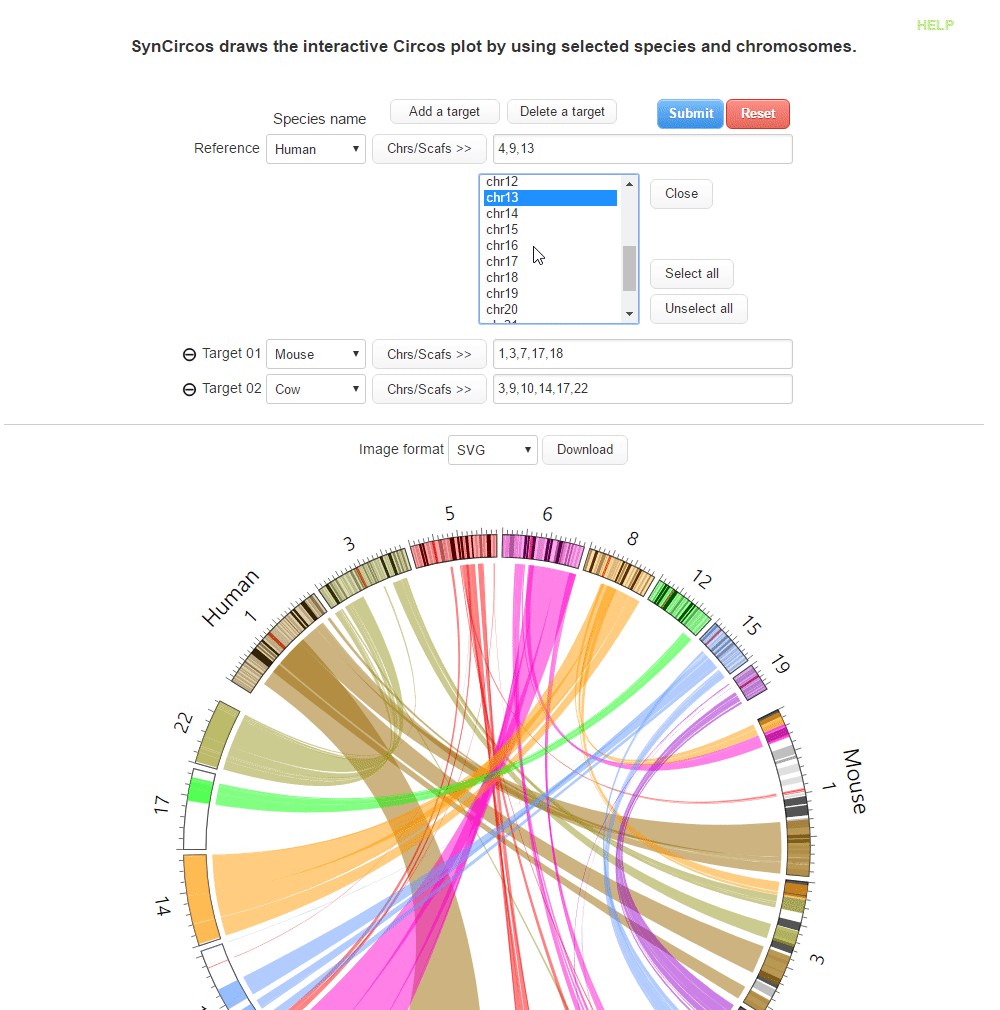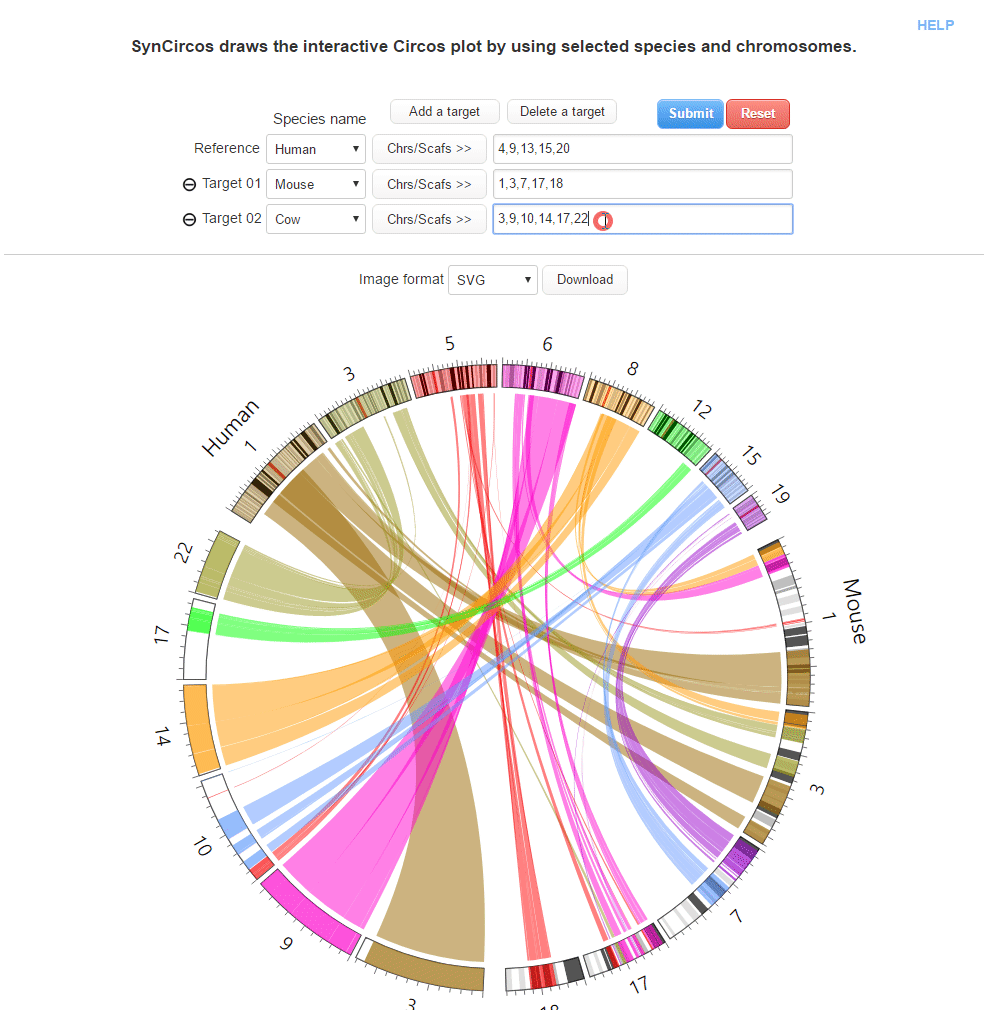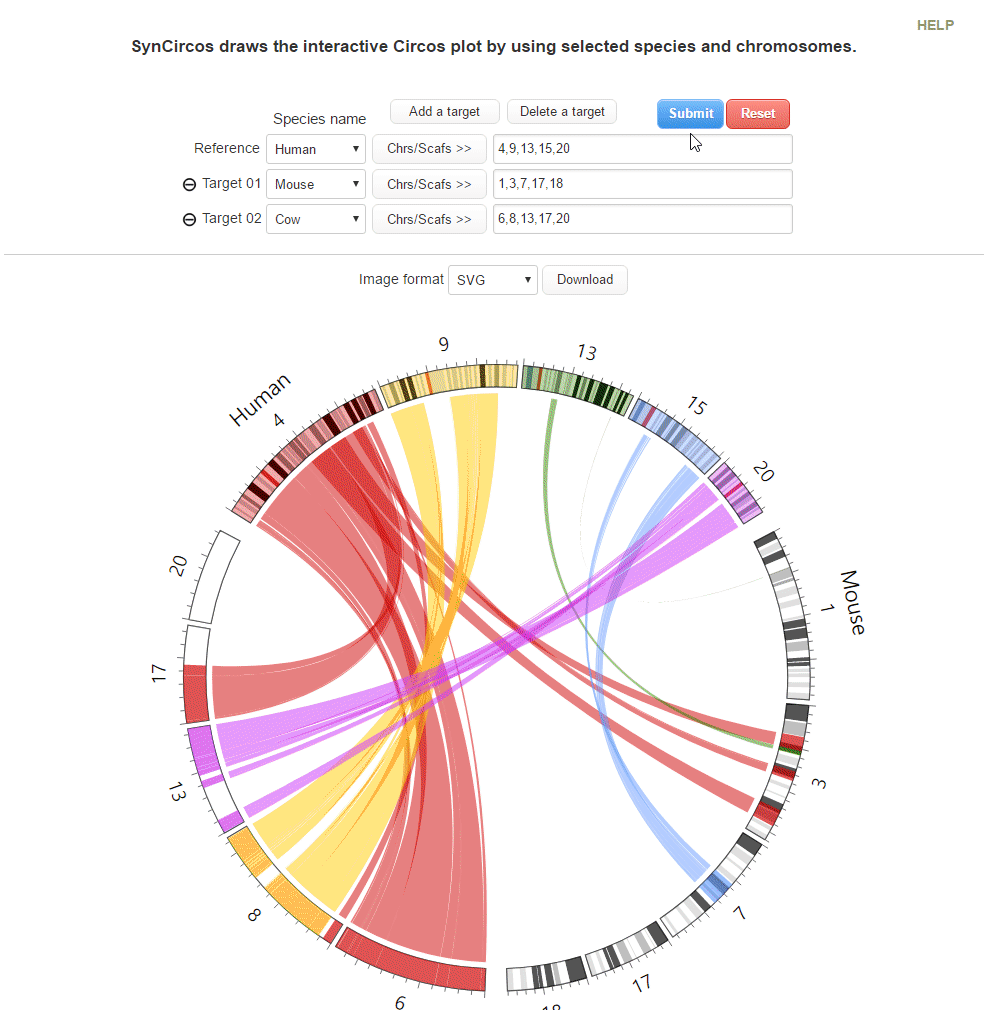 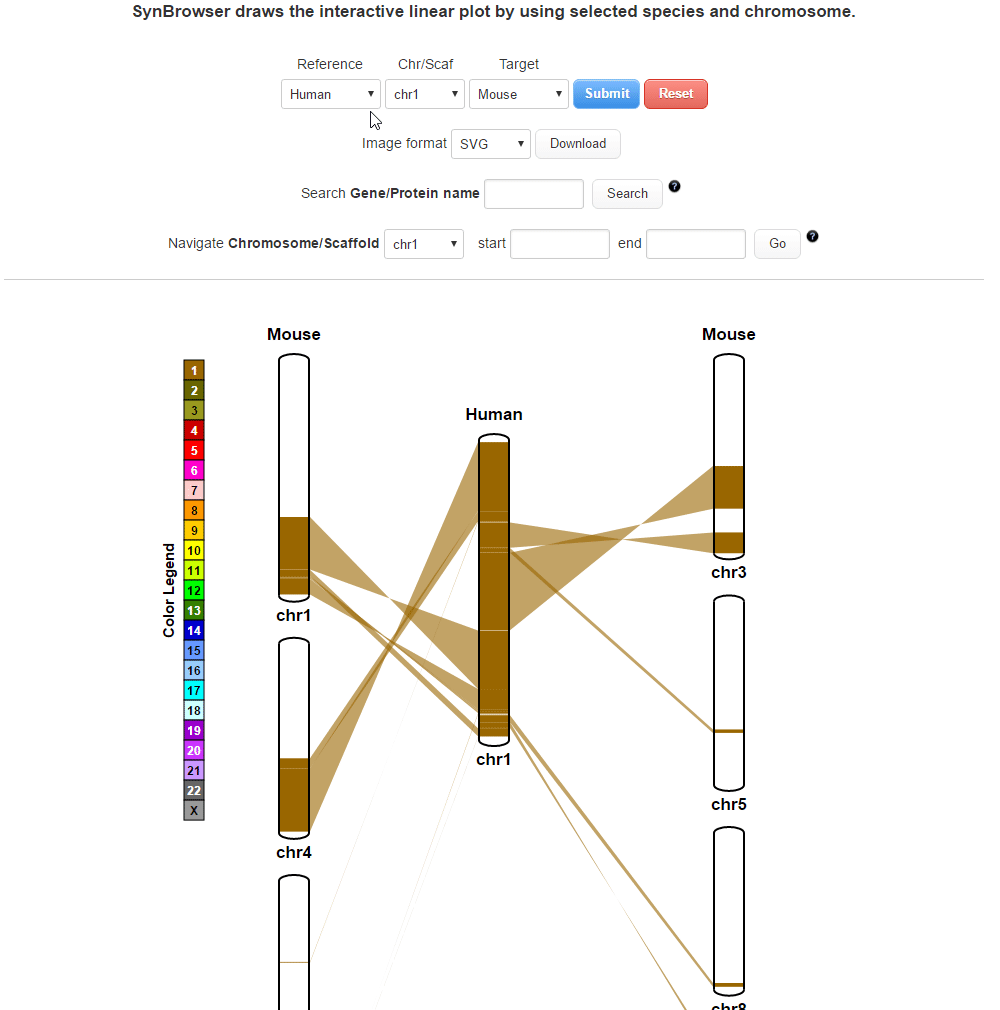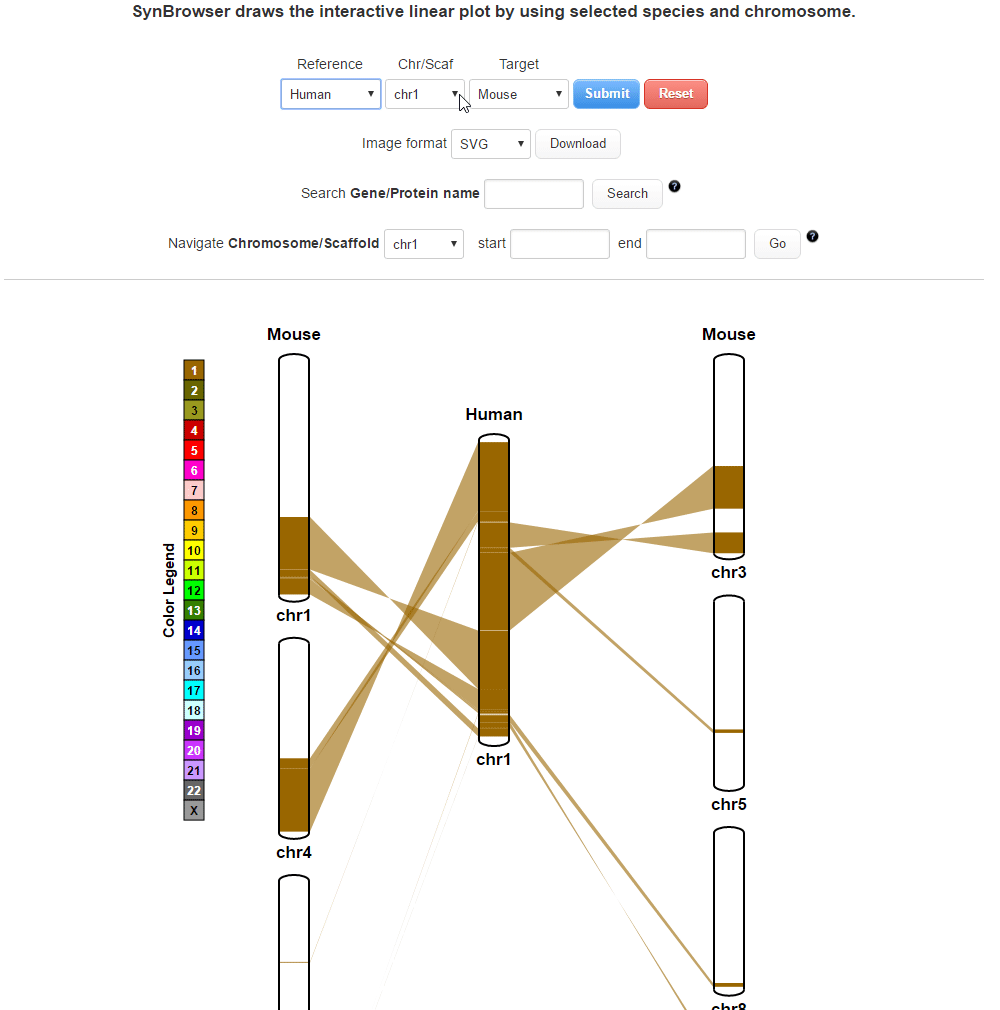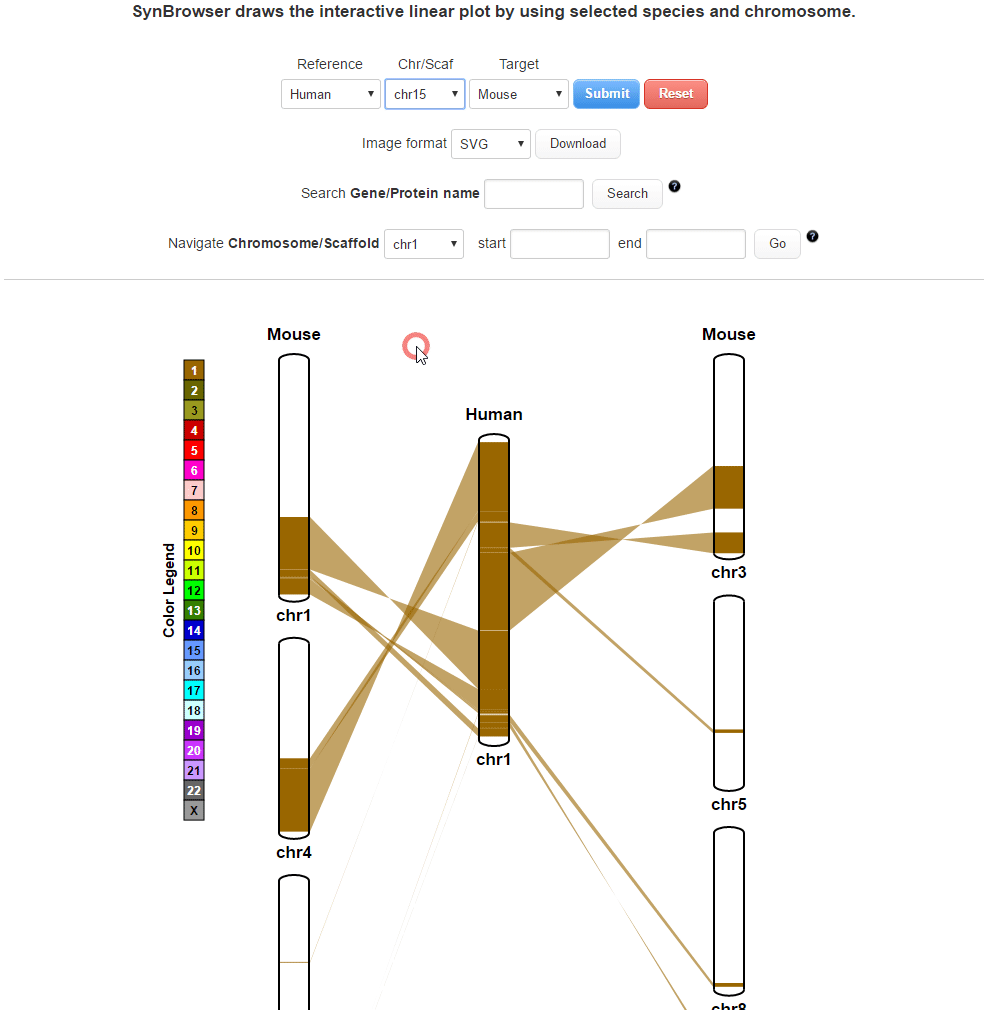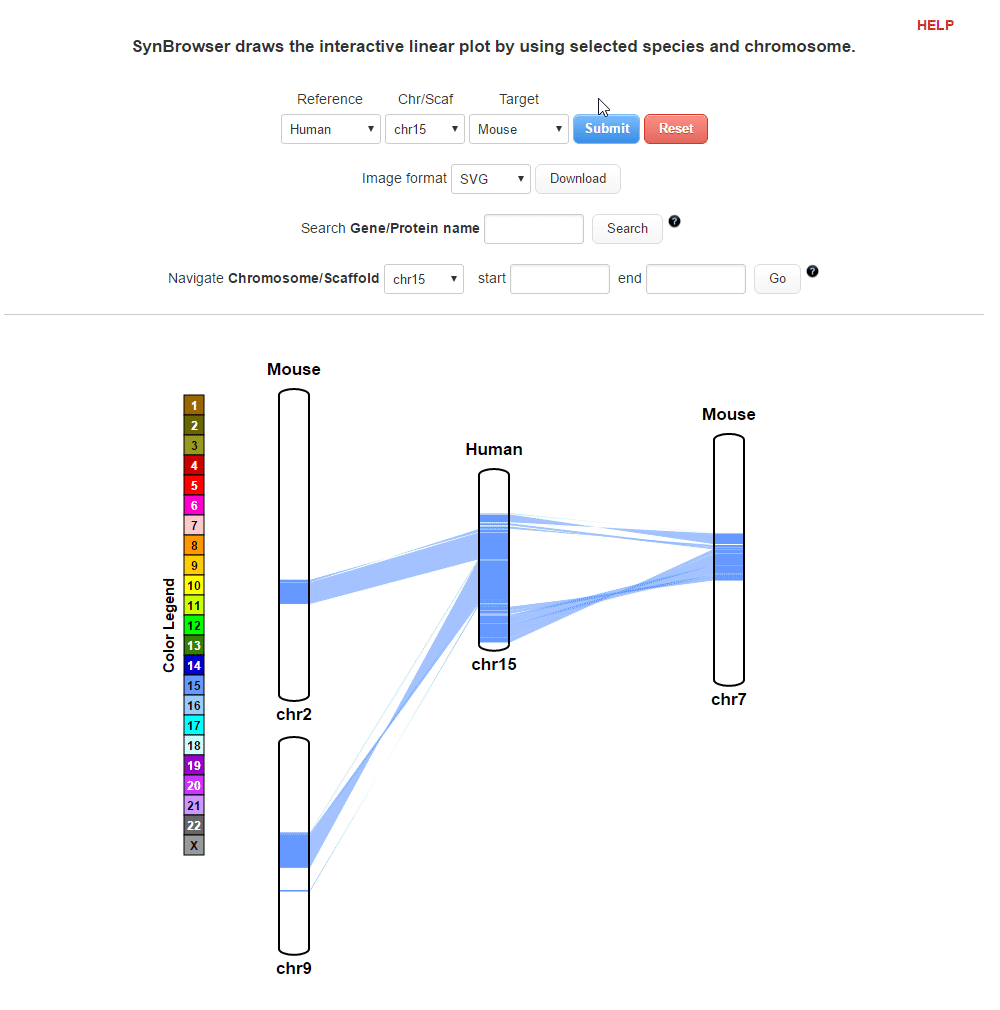
| |
| (2) Publishing a website | |
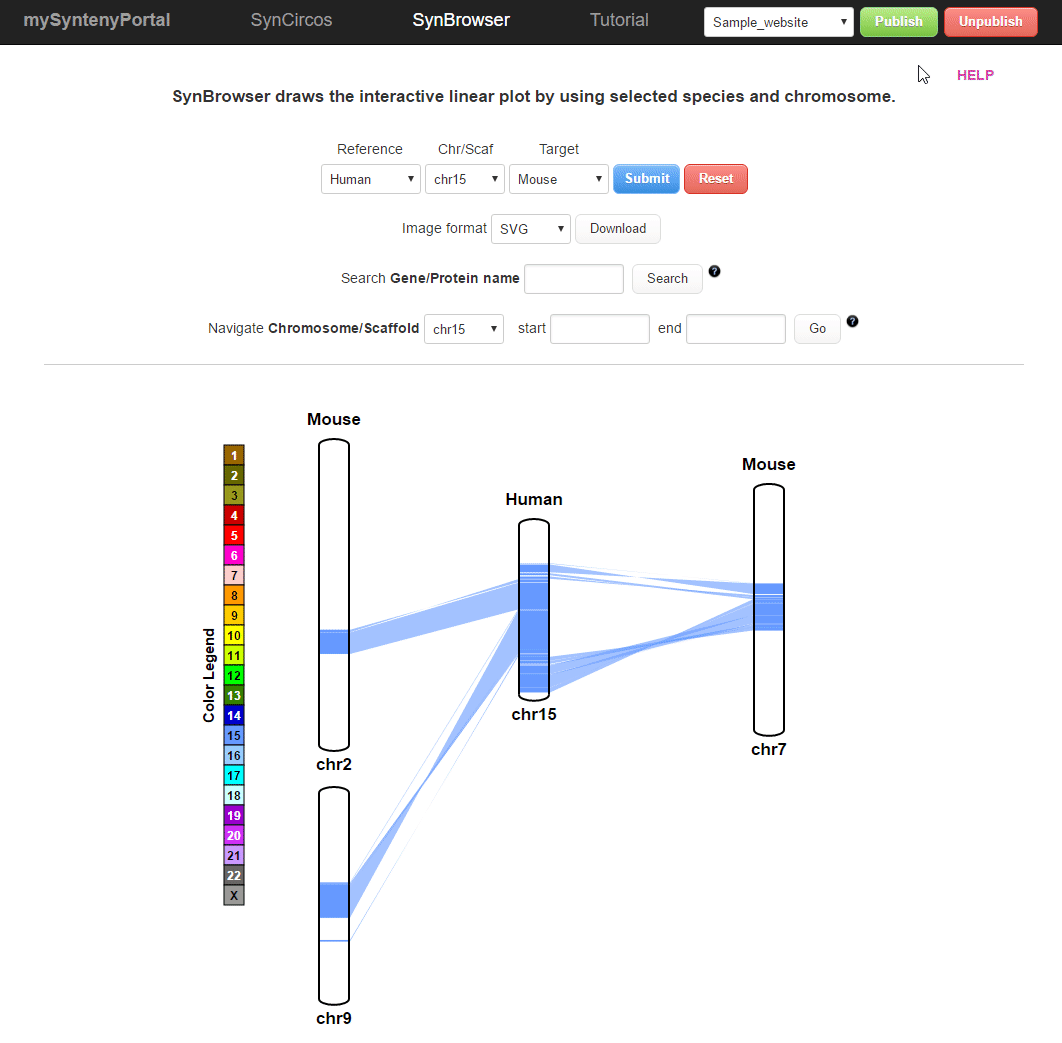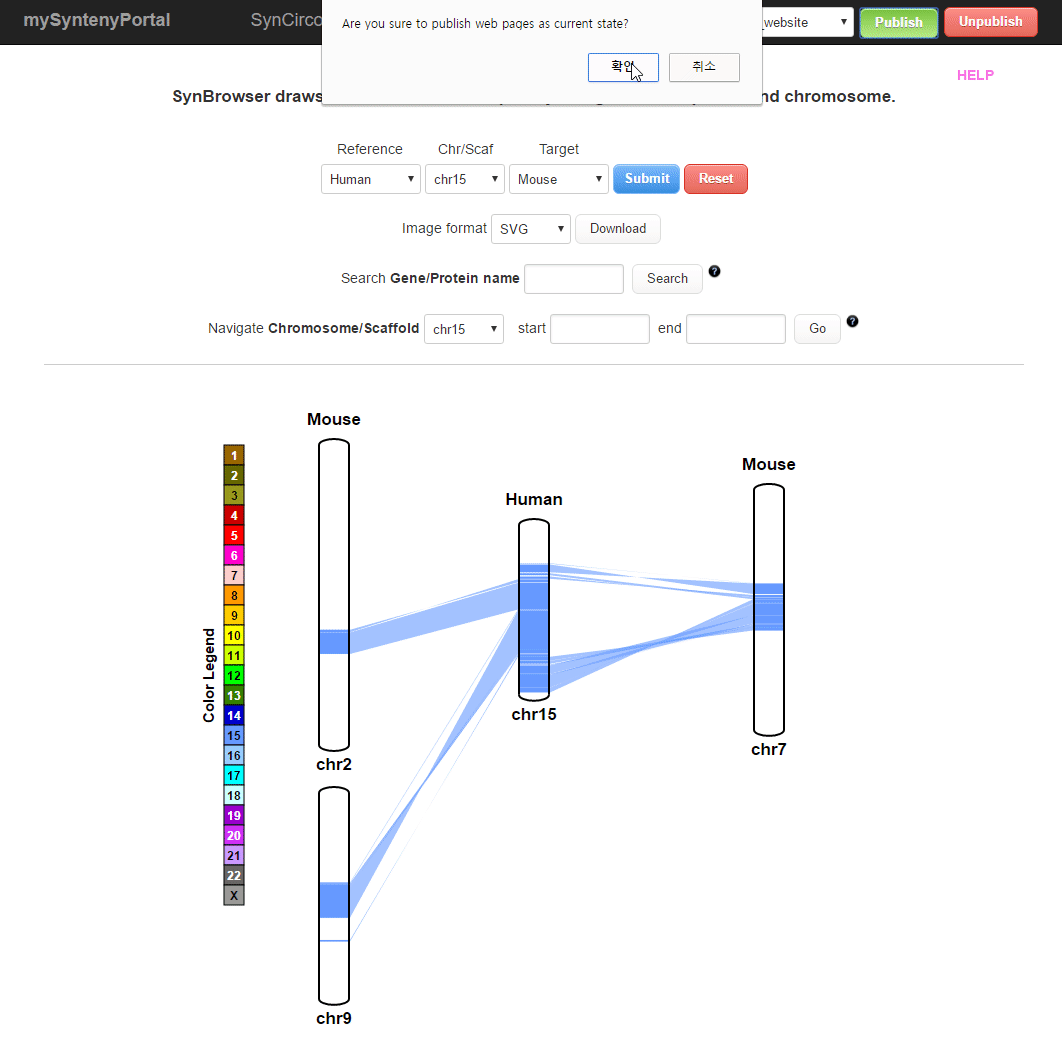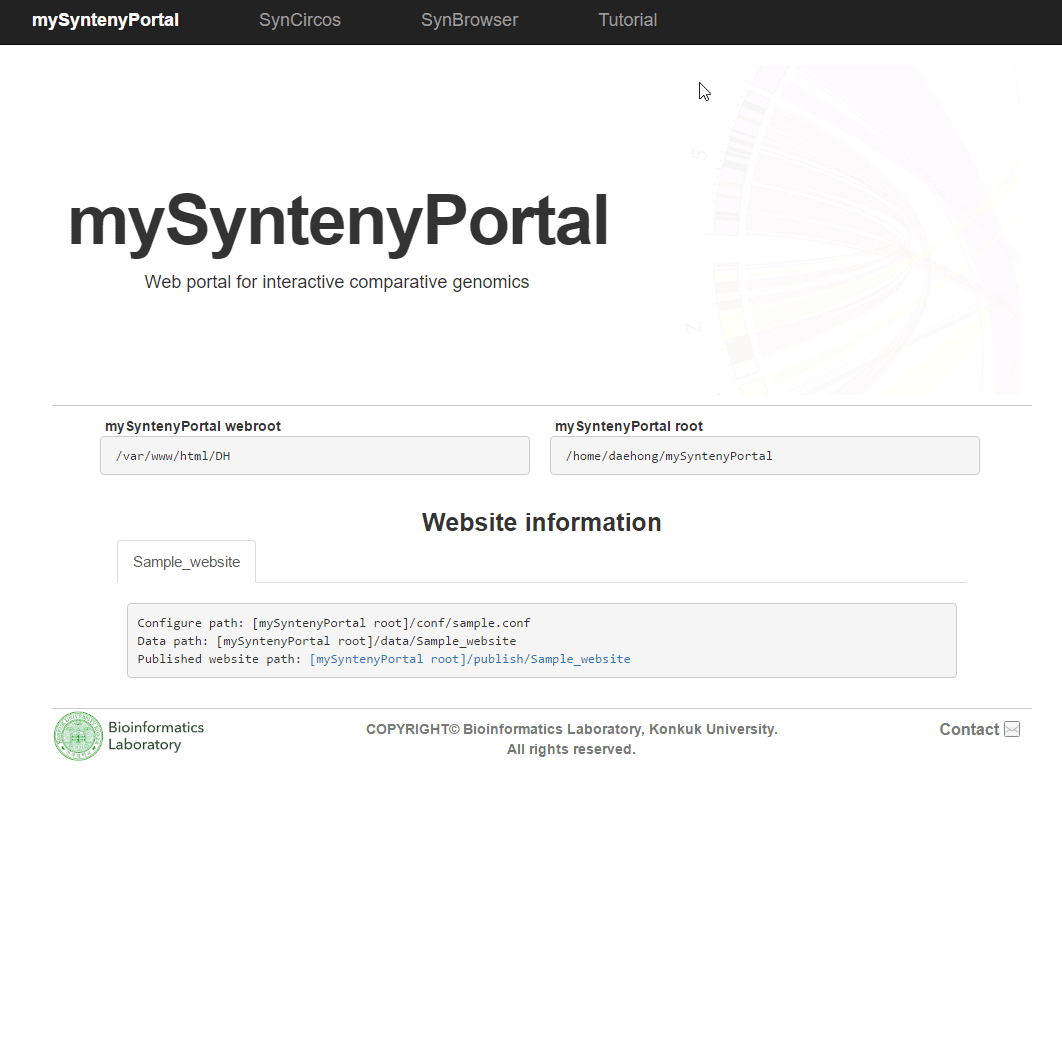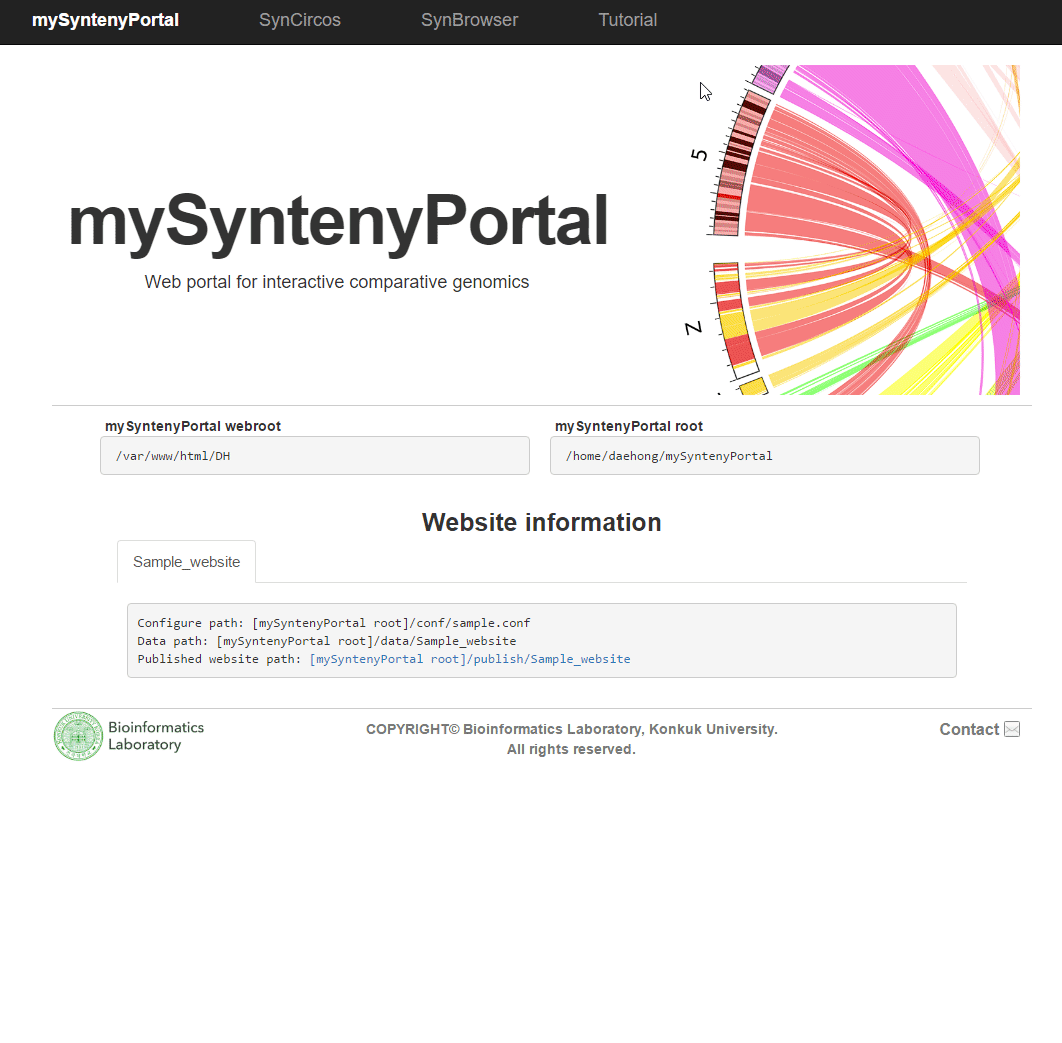 | |
| (3) An example of a pubilshed website | |
 | |
| (4) Published website information | |
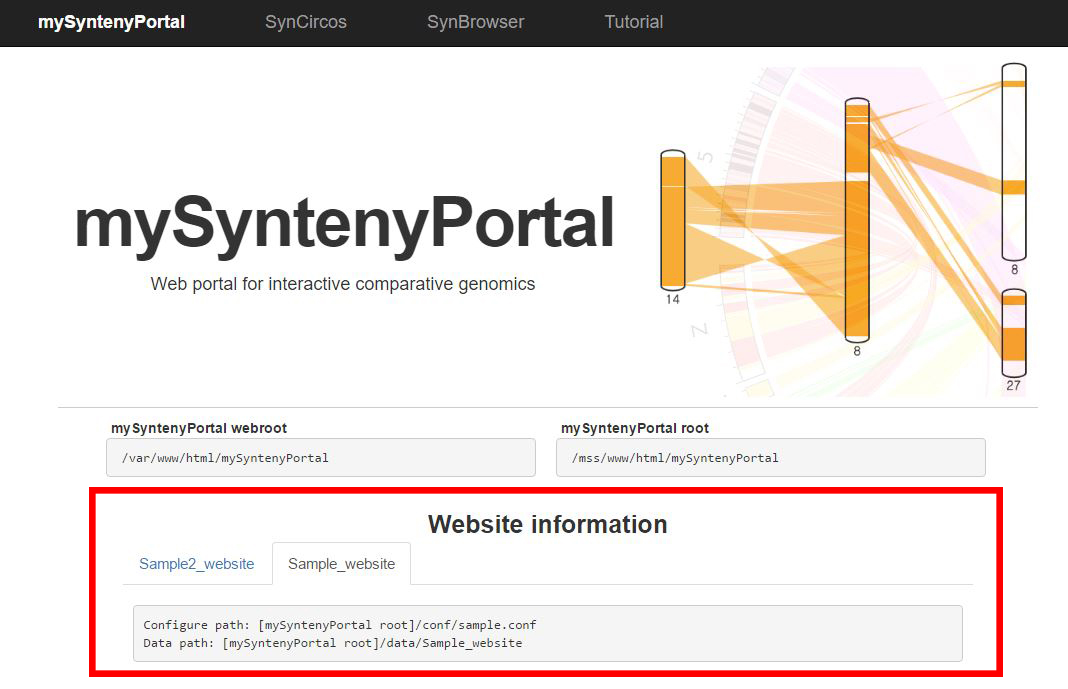
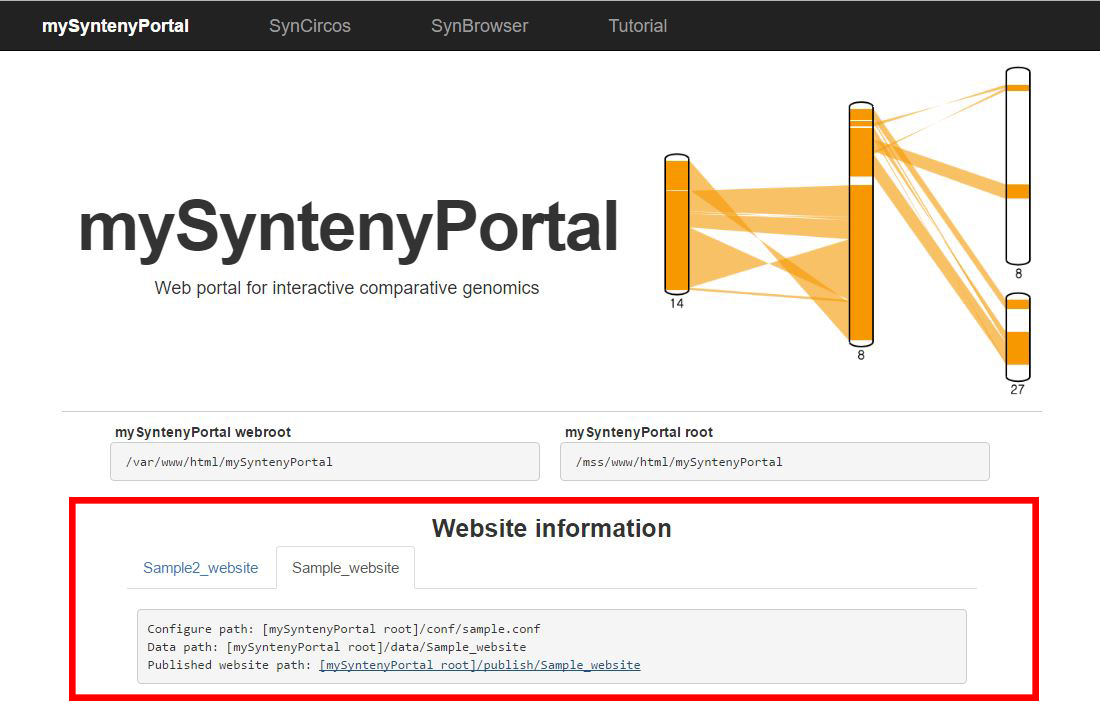
| |
| (5) Unpublishing a selected website | |
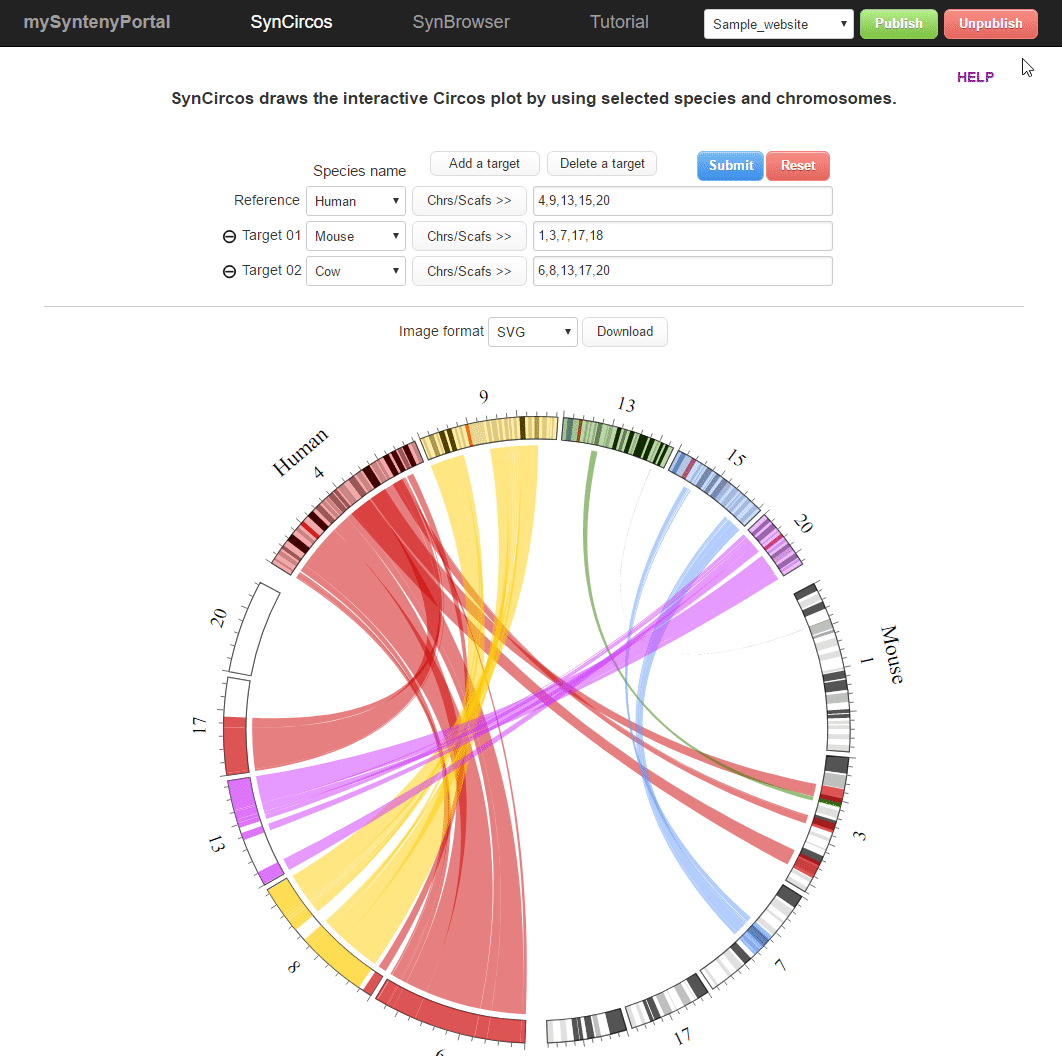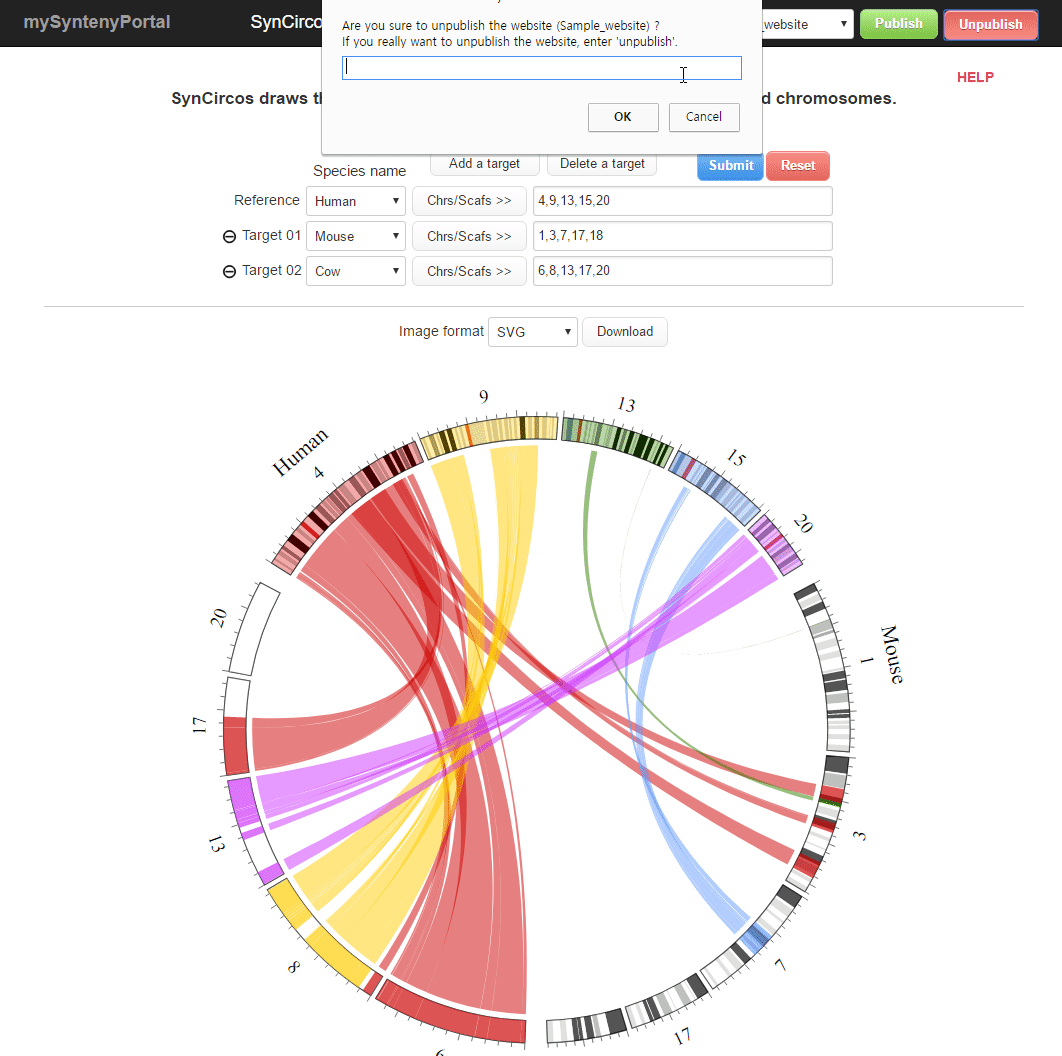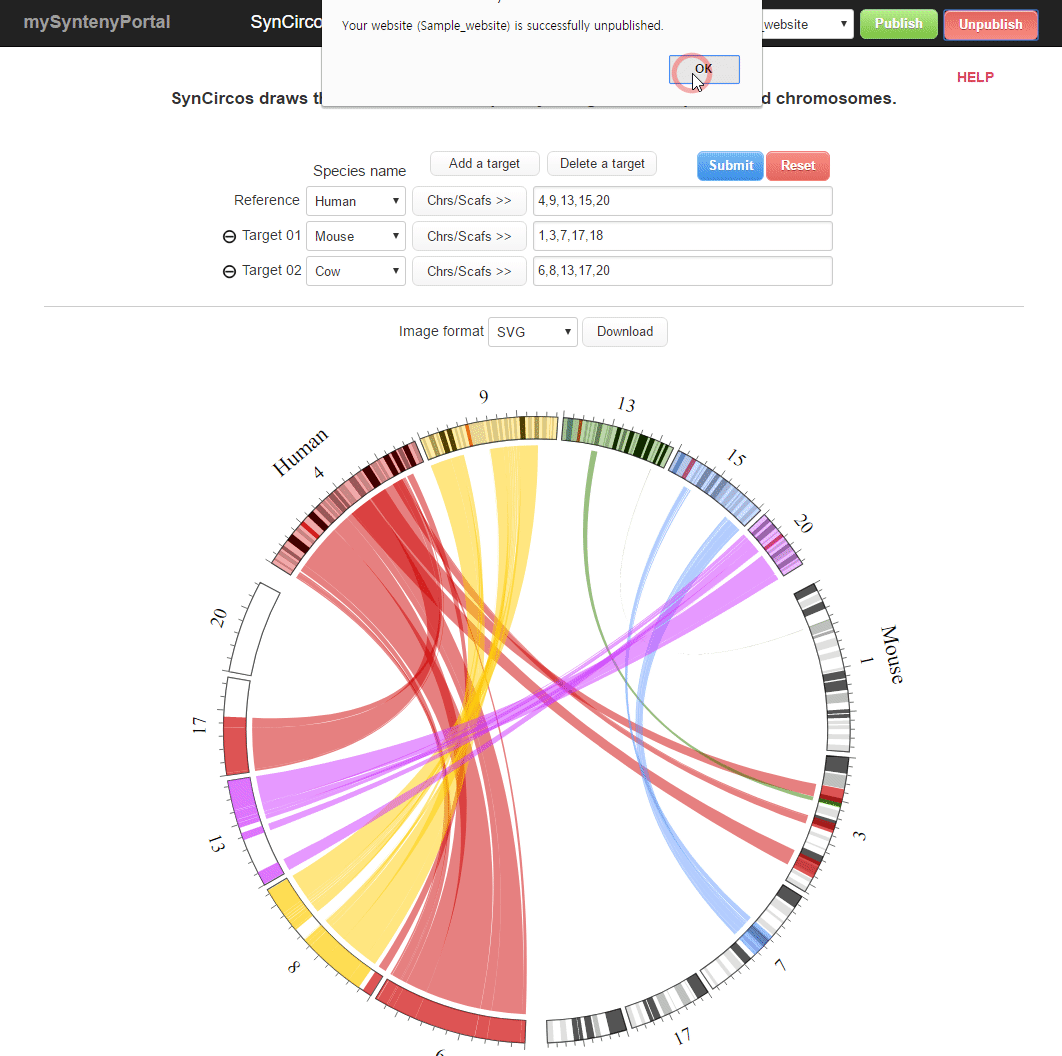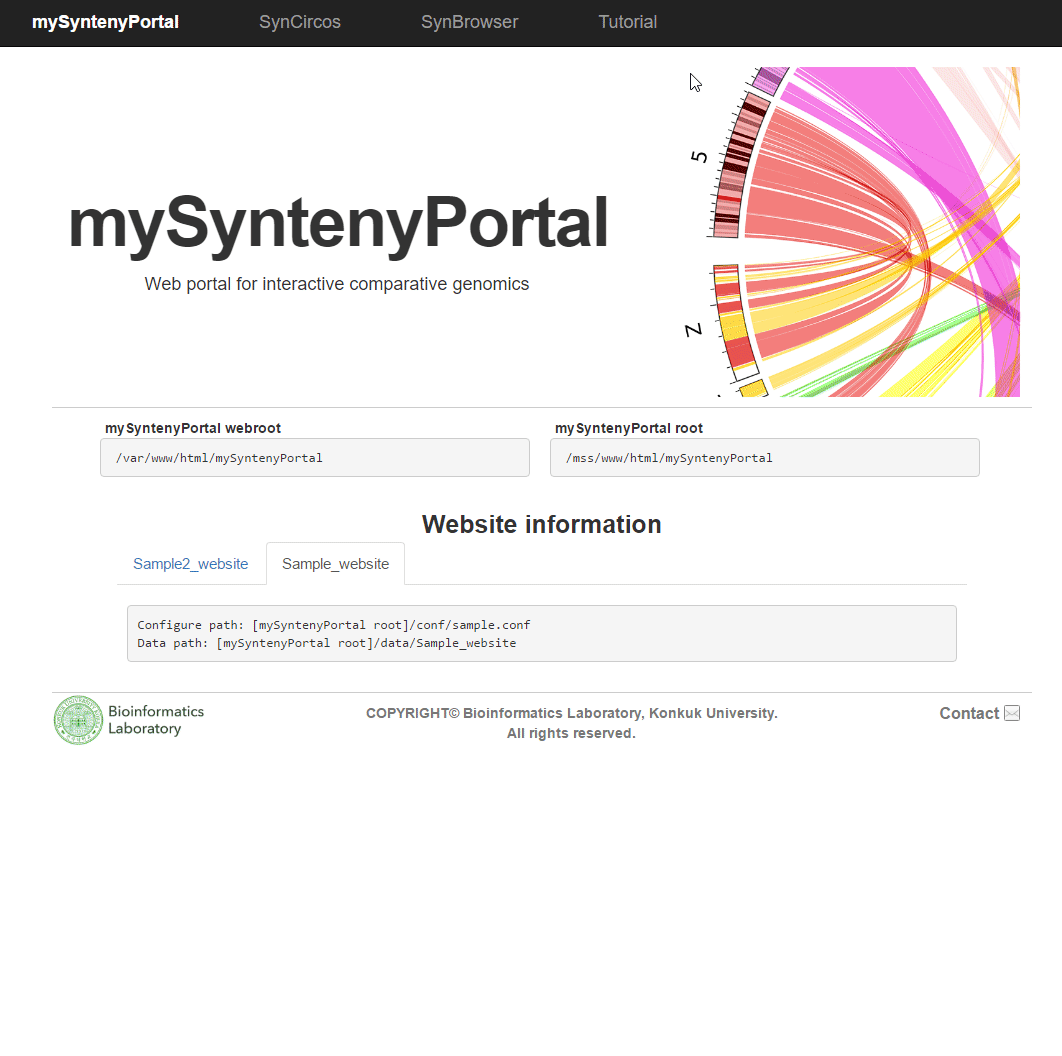 | |
SynCircos
SynCircos draws the interactive Circos plot by using selected species and chormosomes.| 1. Drawing a plot | |
|---|---|
| (1) Selecting a reference species and chromosomes (or scaffolds) | |
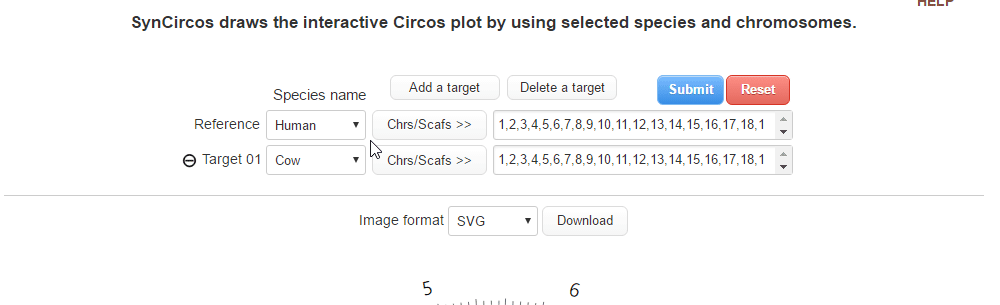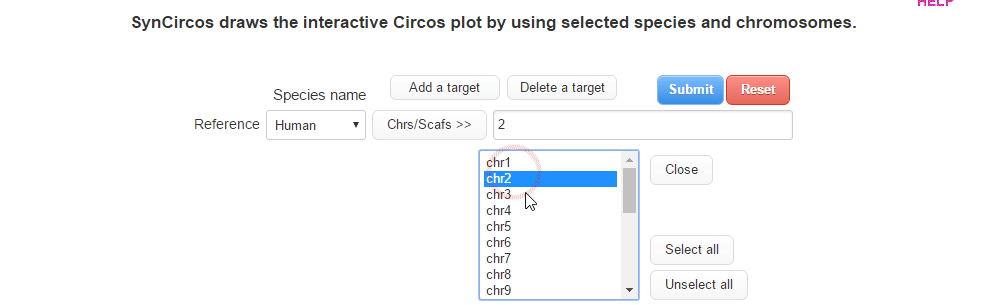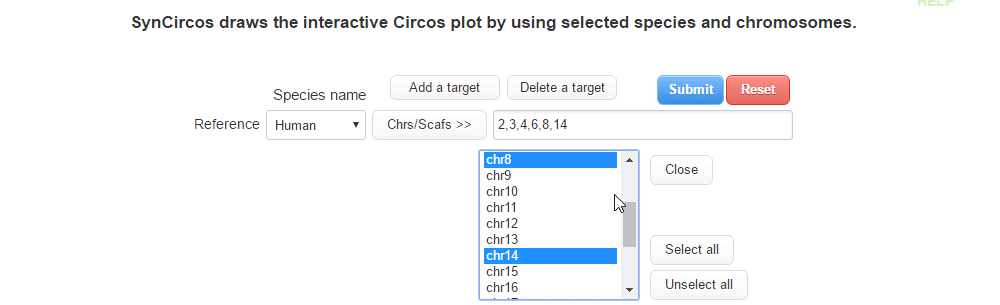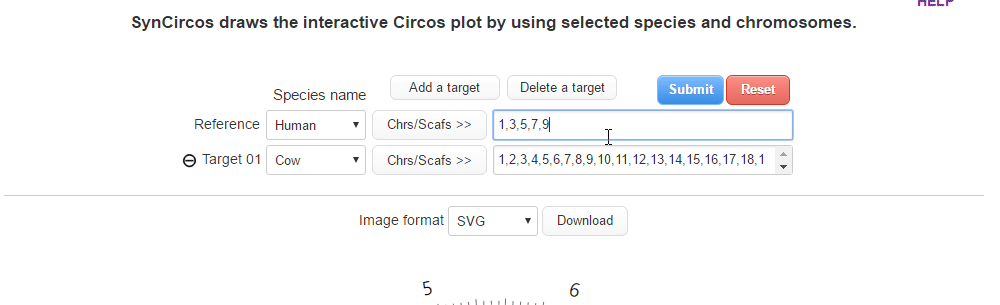 | |
| (2) Selecing a target species and chromosomes (or scaffolds) | |
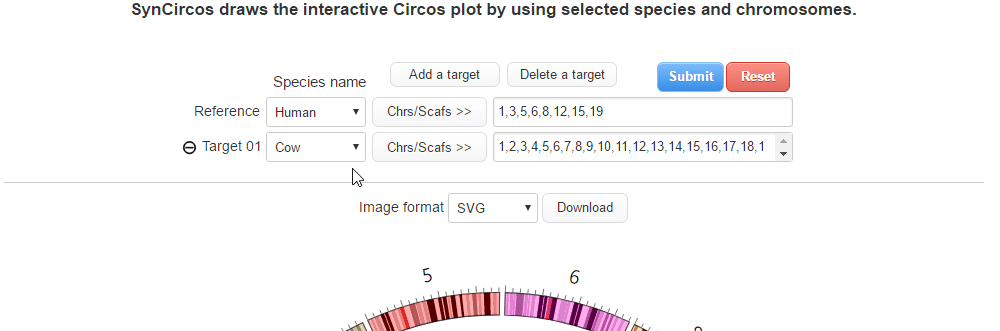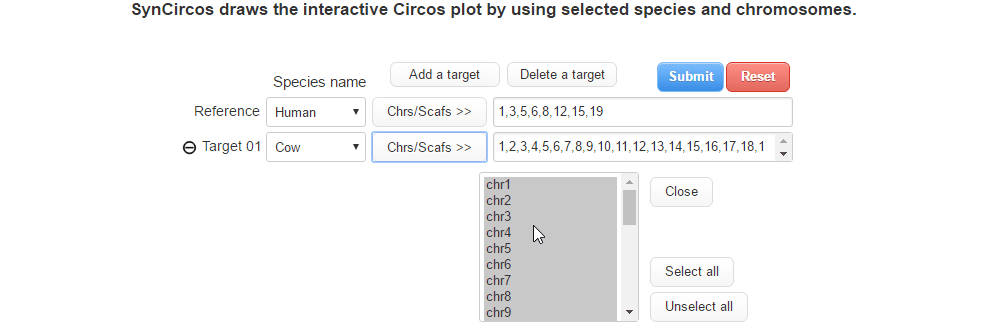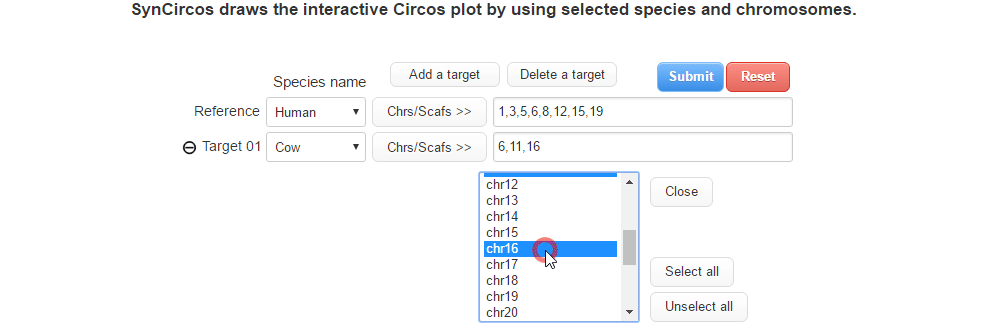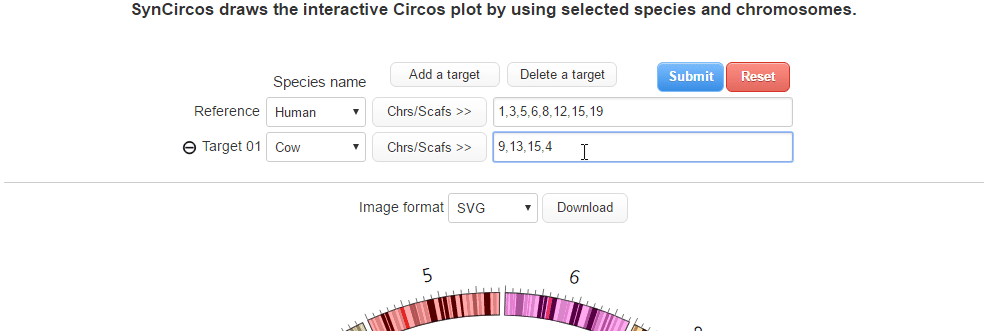 | |
| (2-1) Adding a target species | |
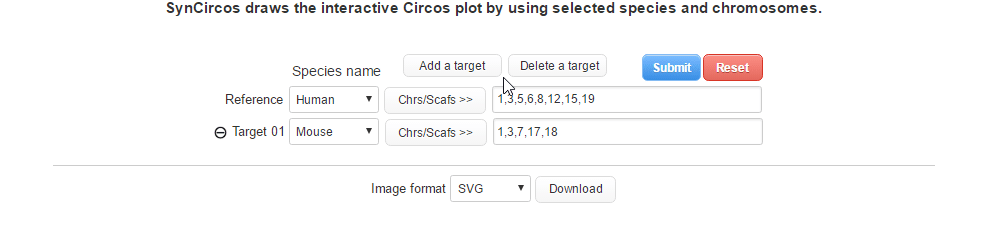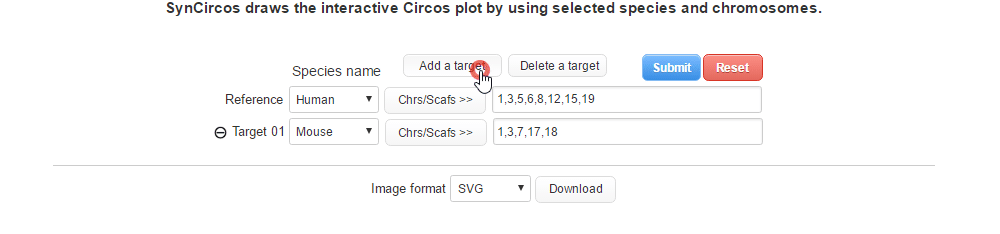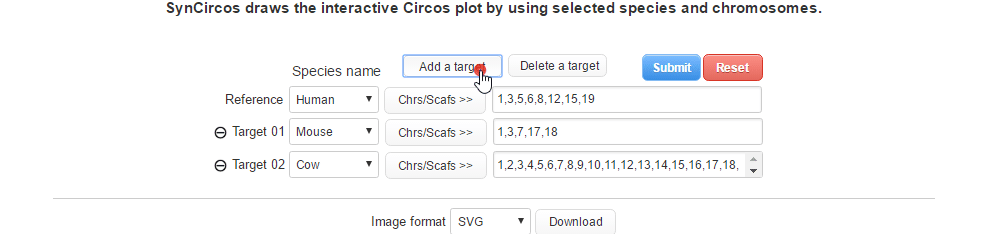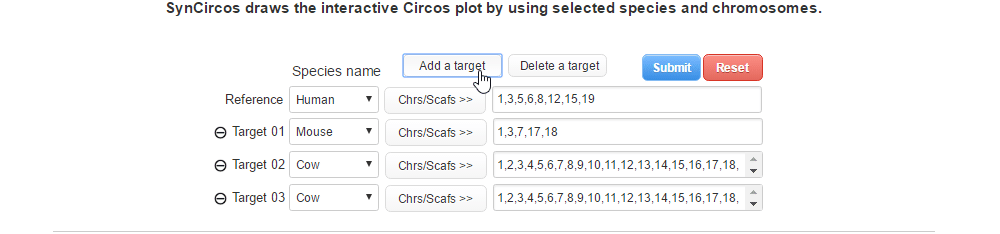 | |
| (2-2) Removing a target species | |
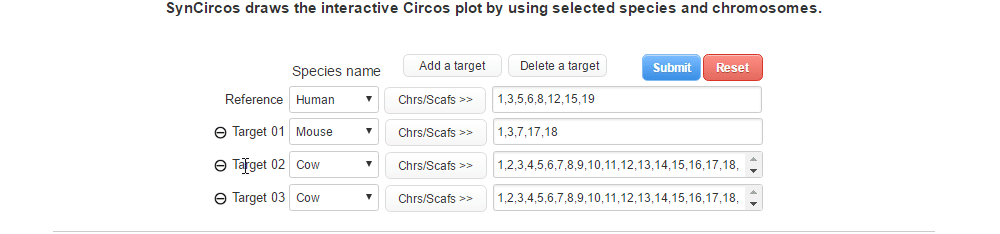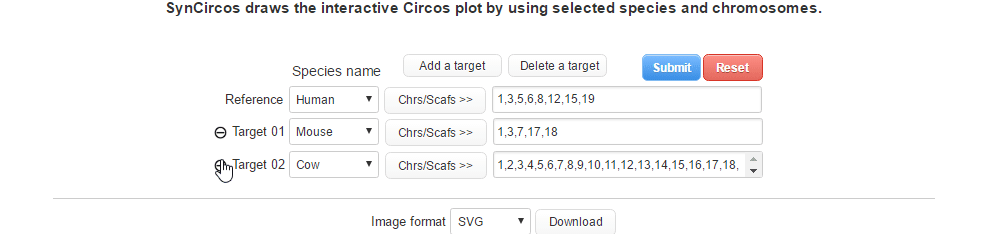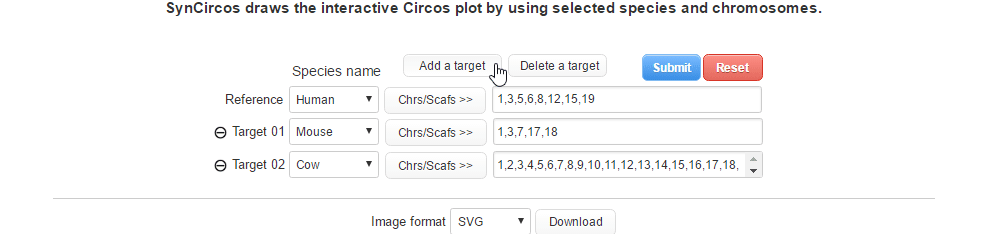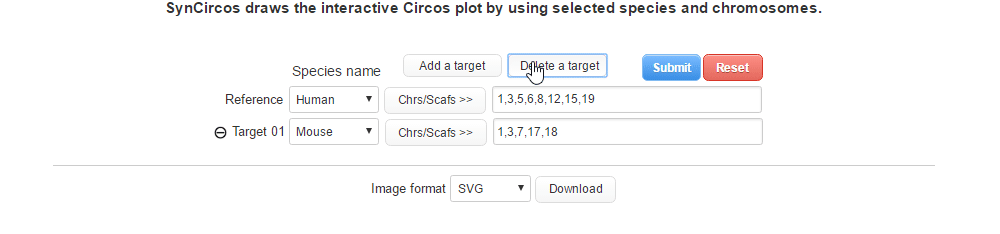 | |
| (3) Selecting a resolution of synteny blocks (Only assemblies input) | |
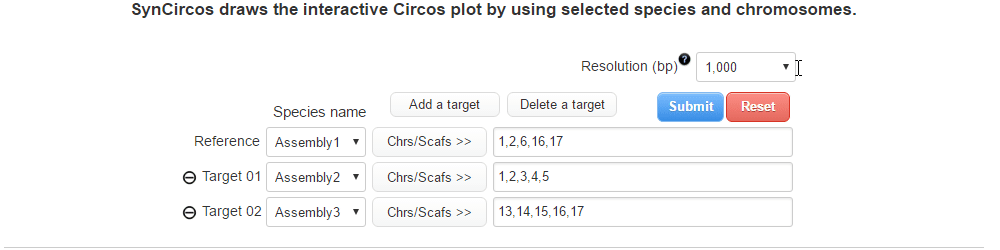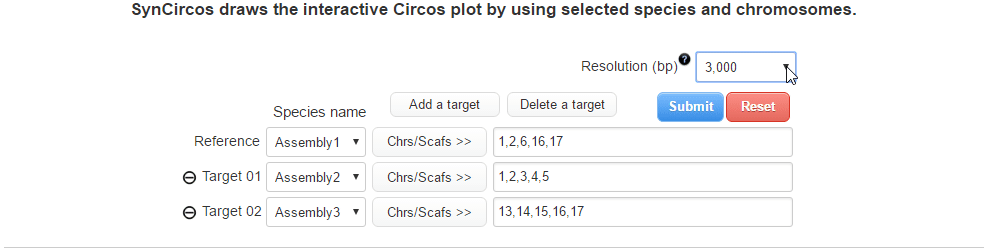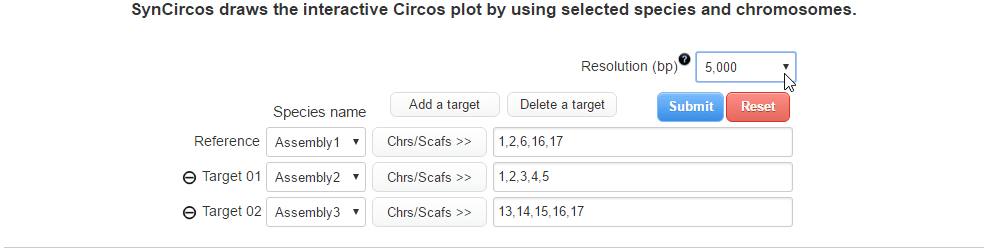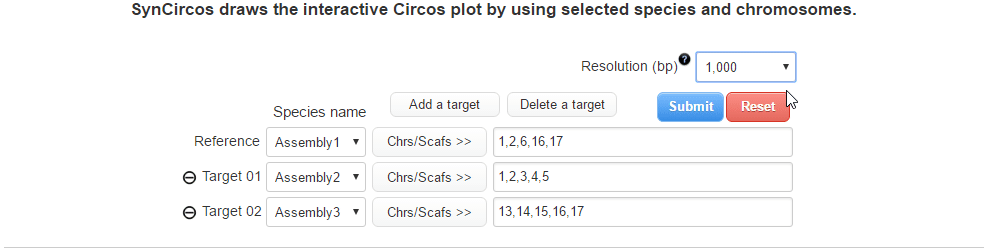 | |
| (4) Clicking 'Submit' button to draw the Circos plot | |
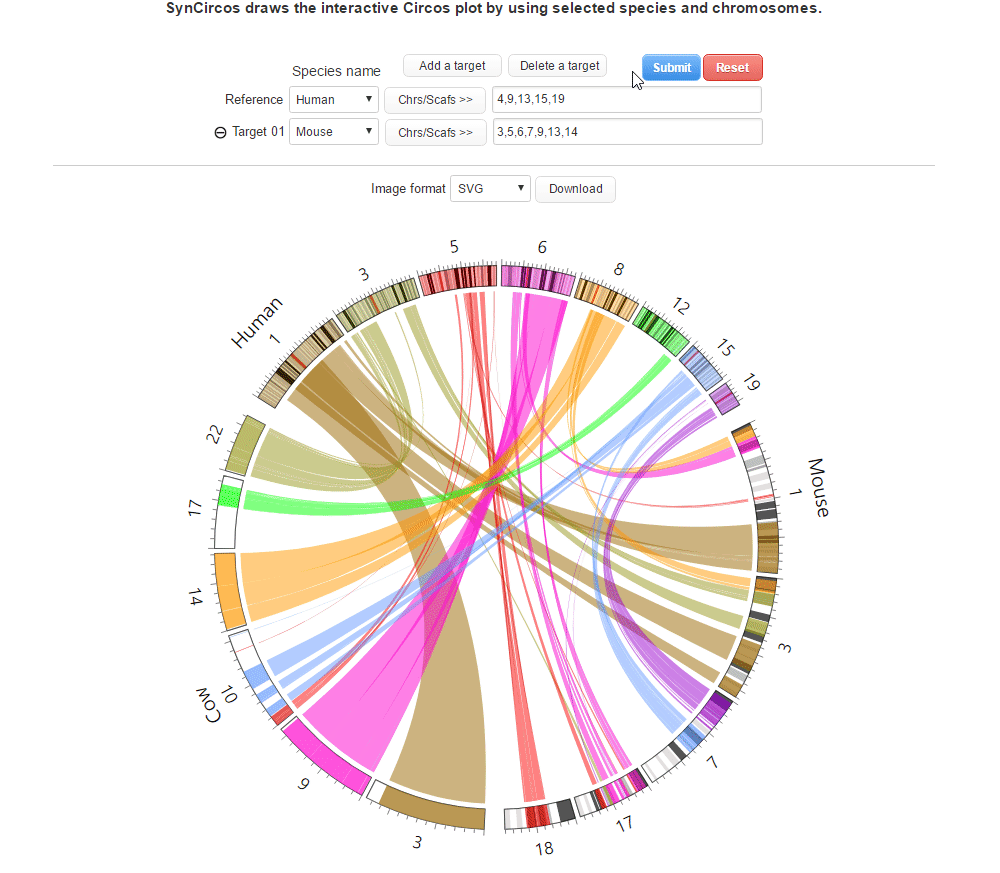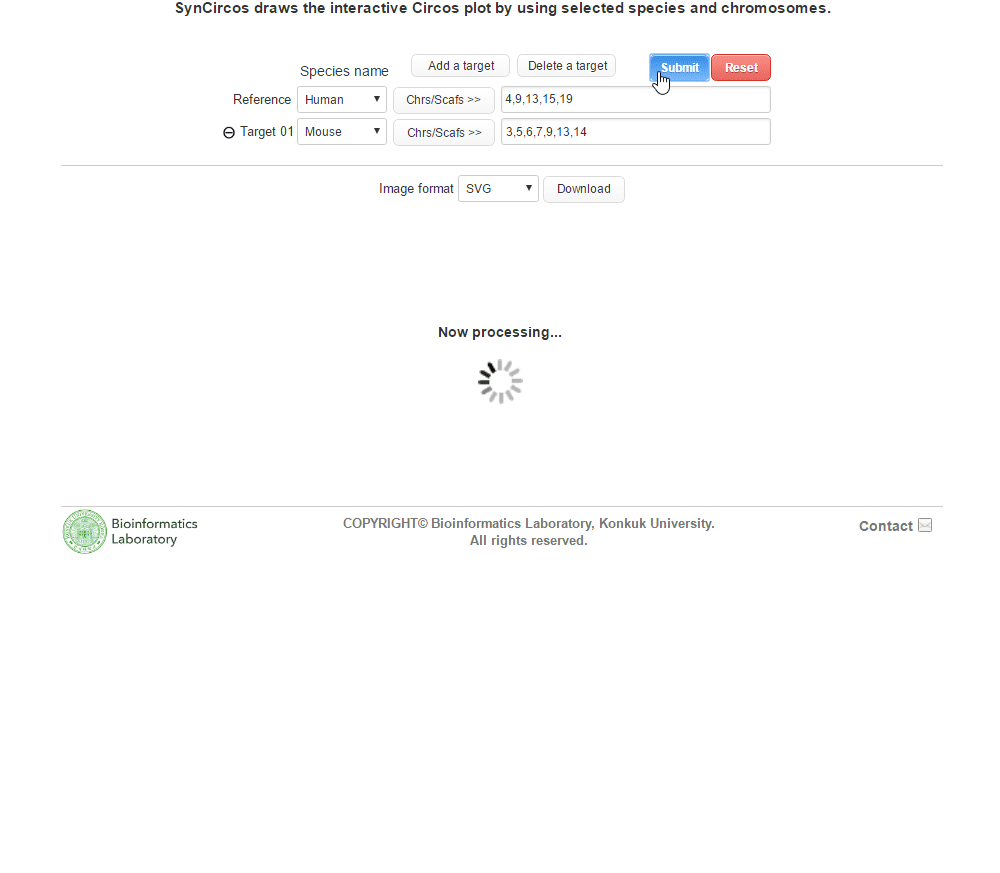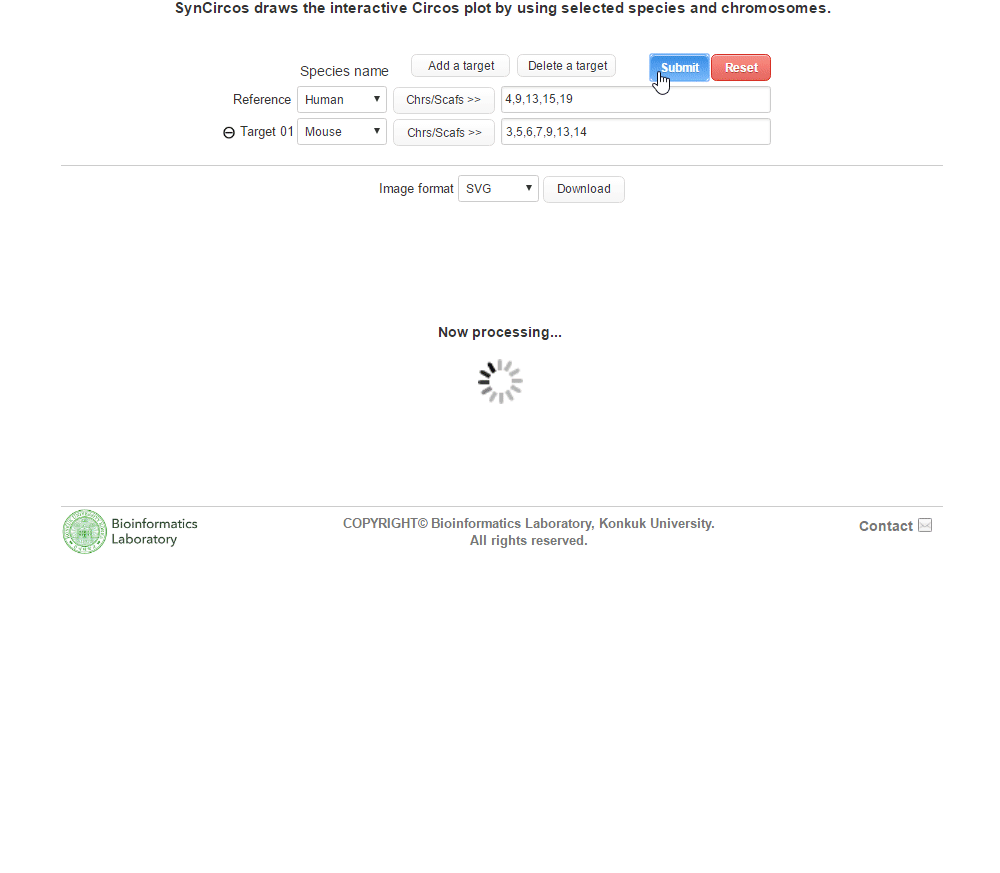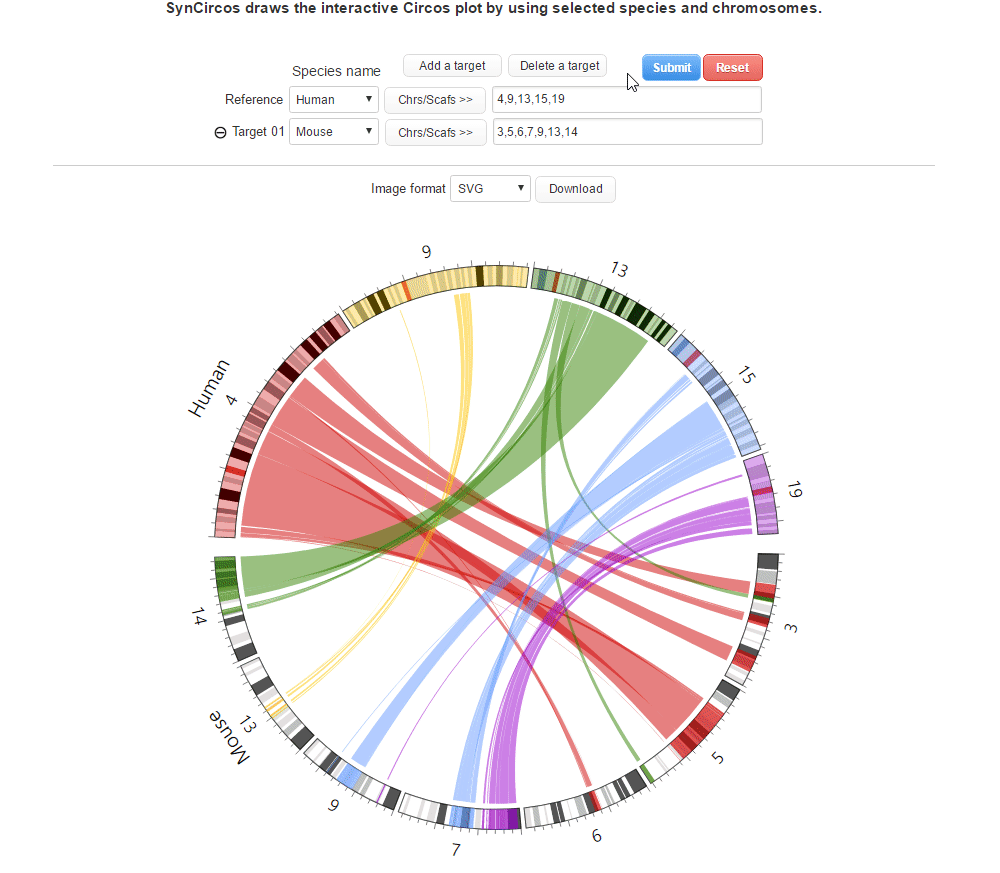 | |
| (5) An example of the Circos plot | |
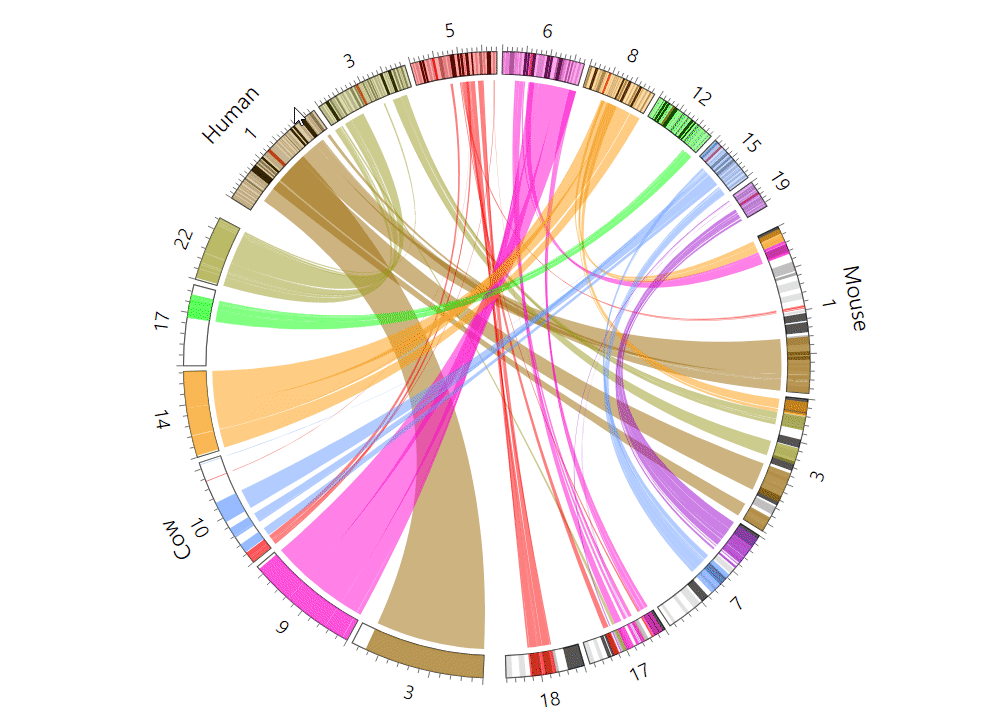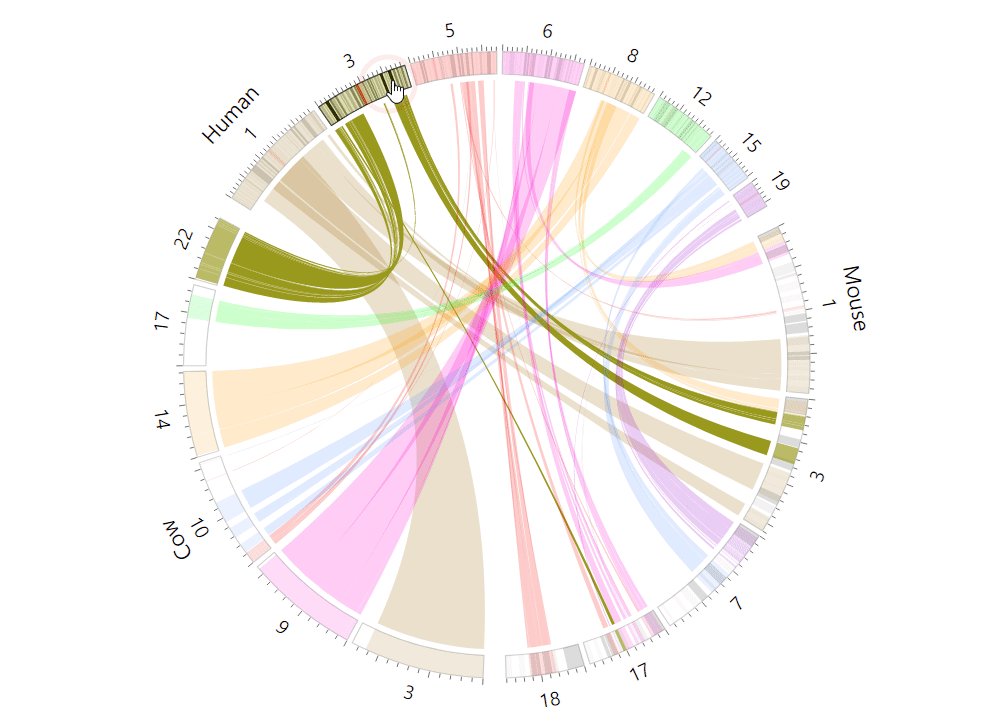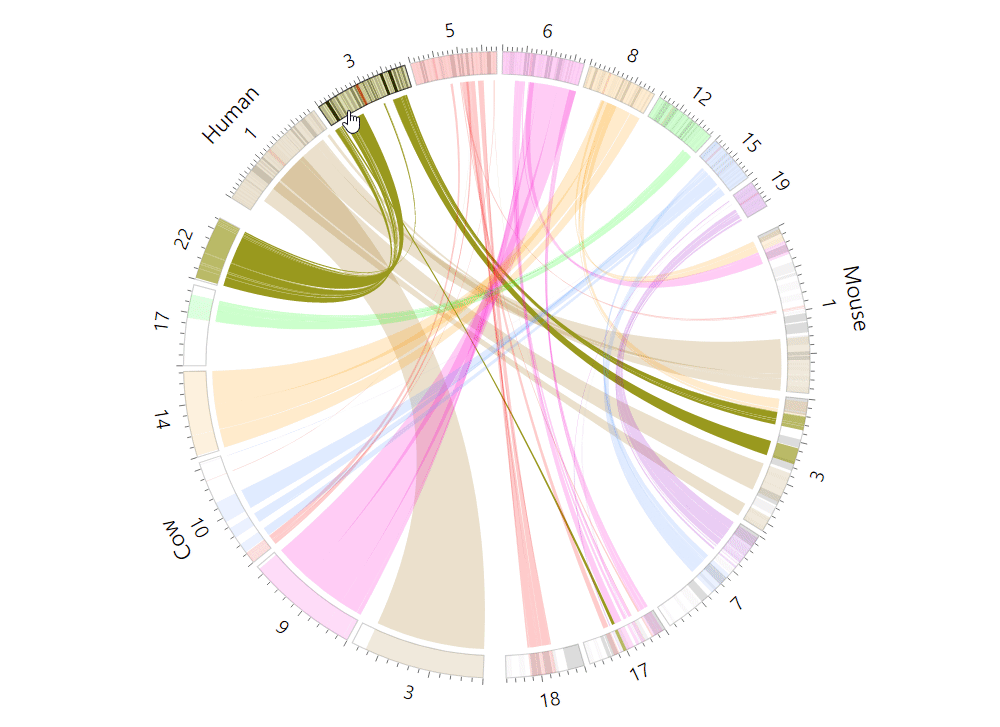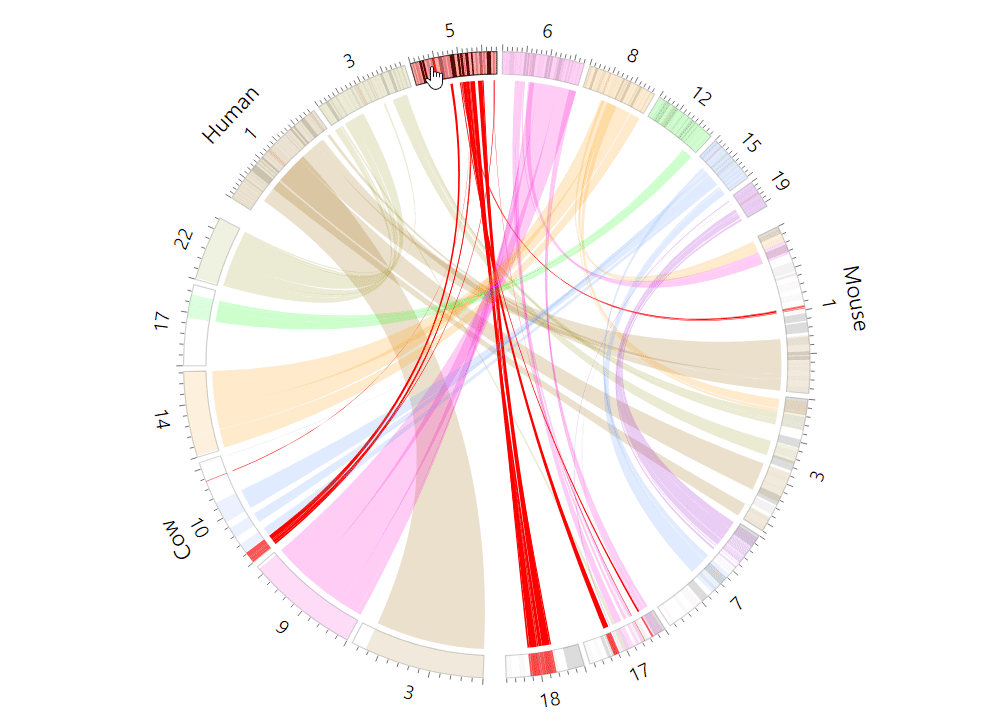 | |
| 2. Downloading a plot | |
|---|---|
| (1) Selecting an image format. | |
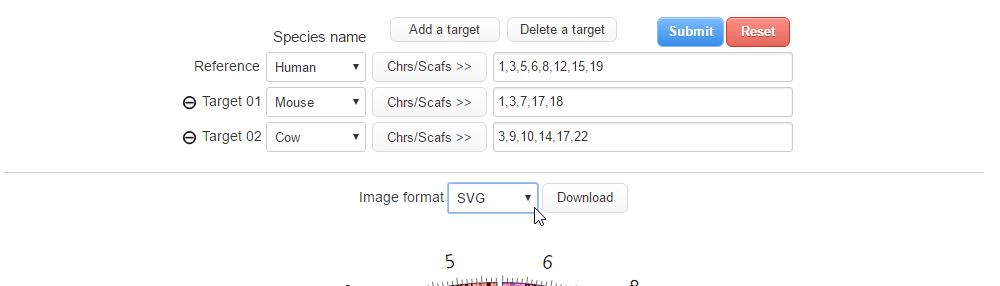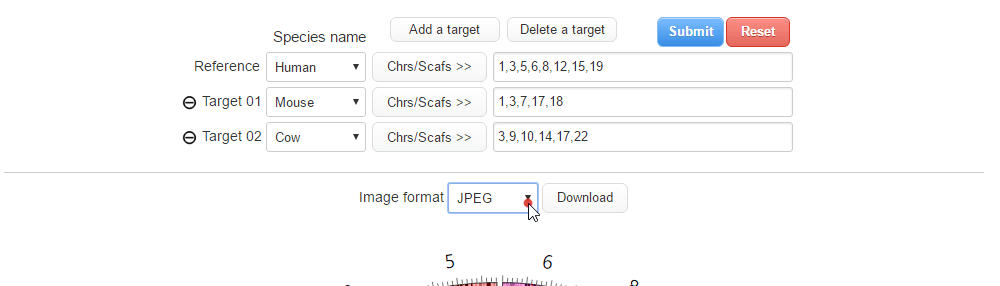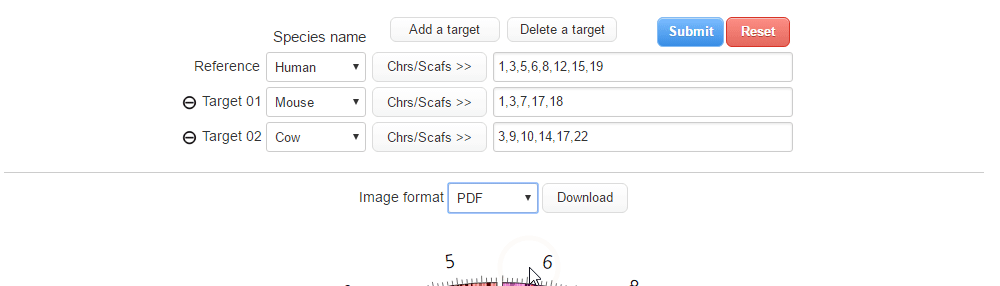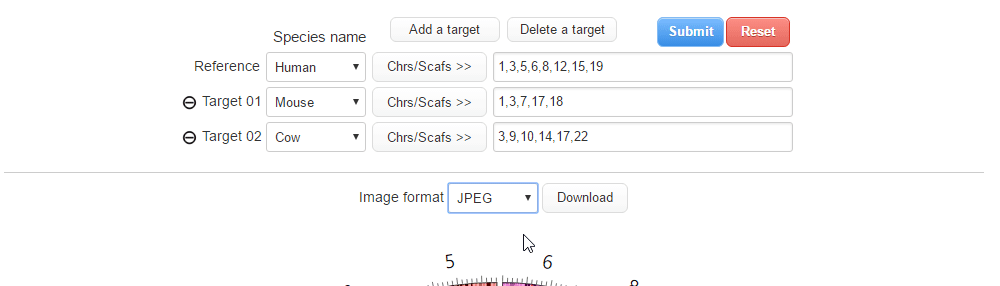 | |
| (2) Clicking on 'Download' button. | |
 | |
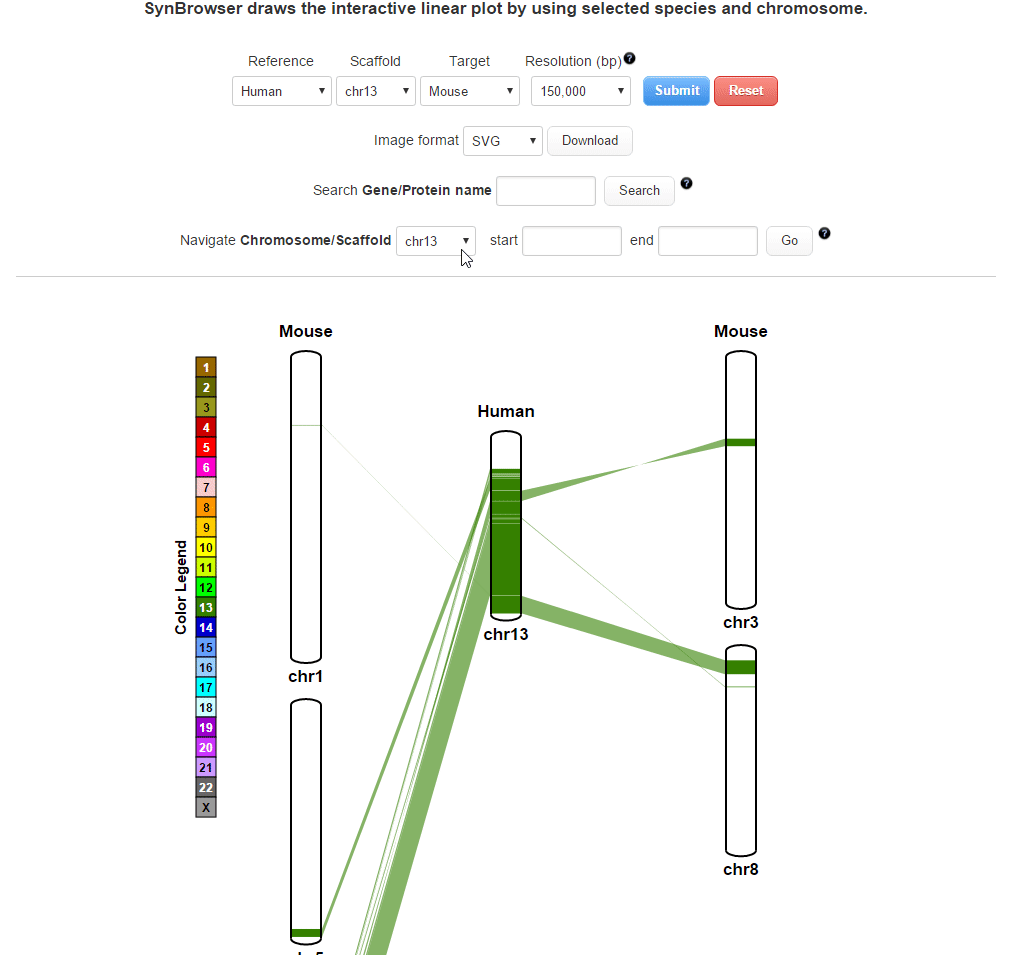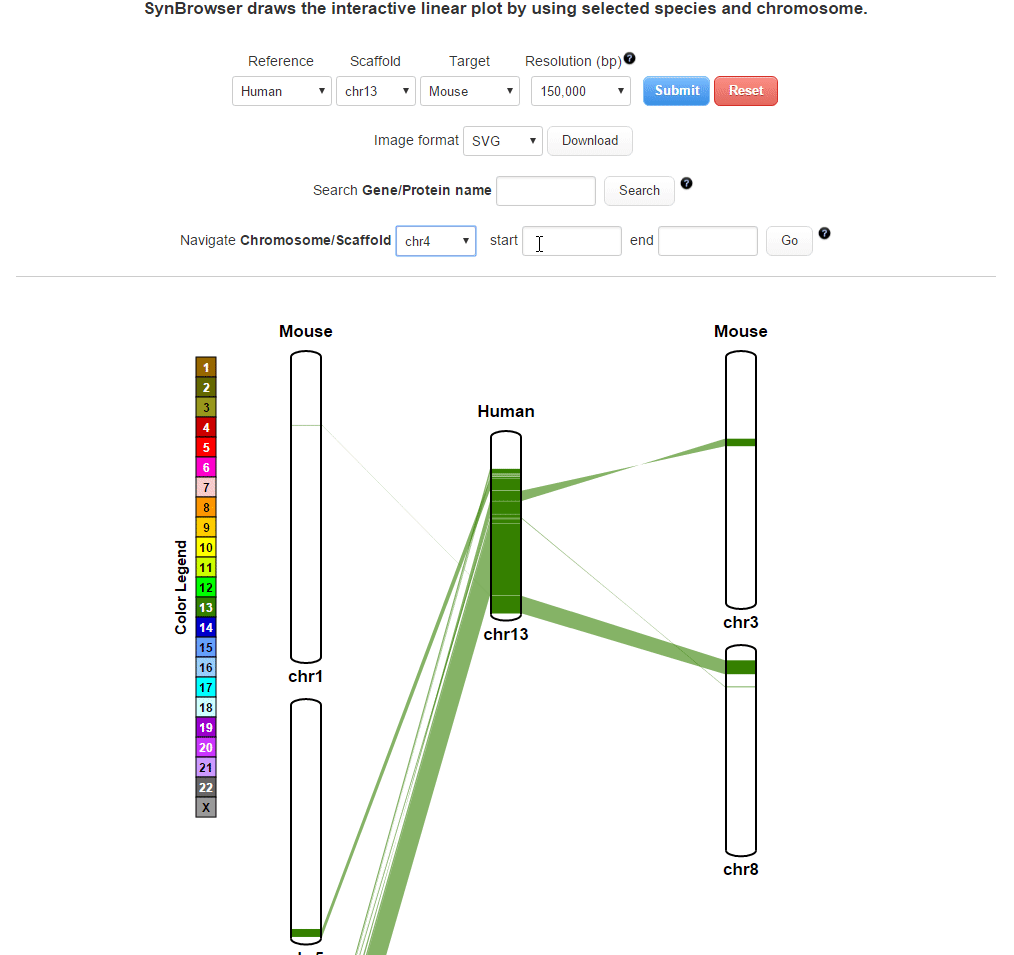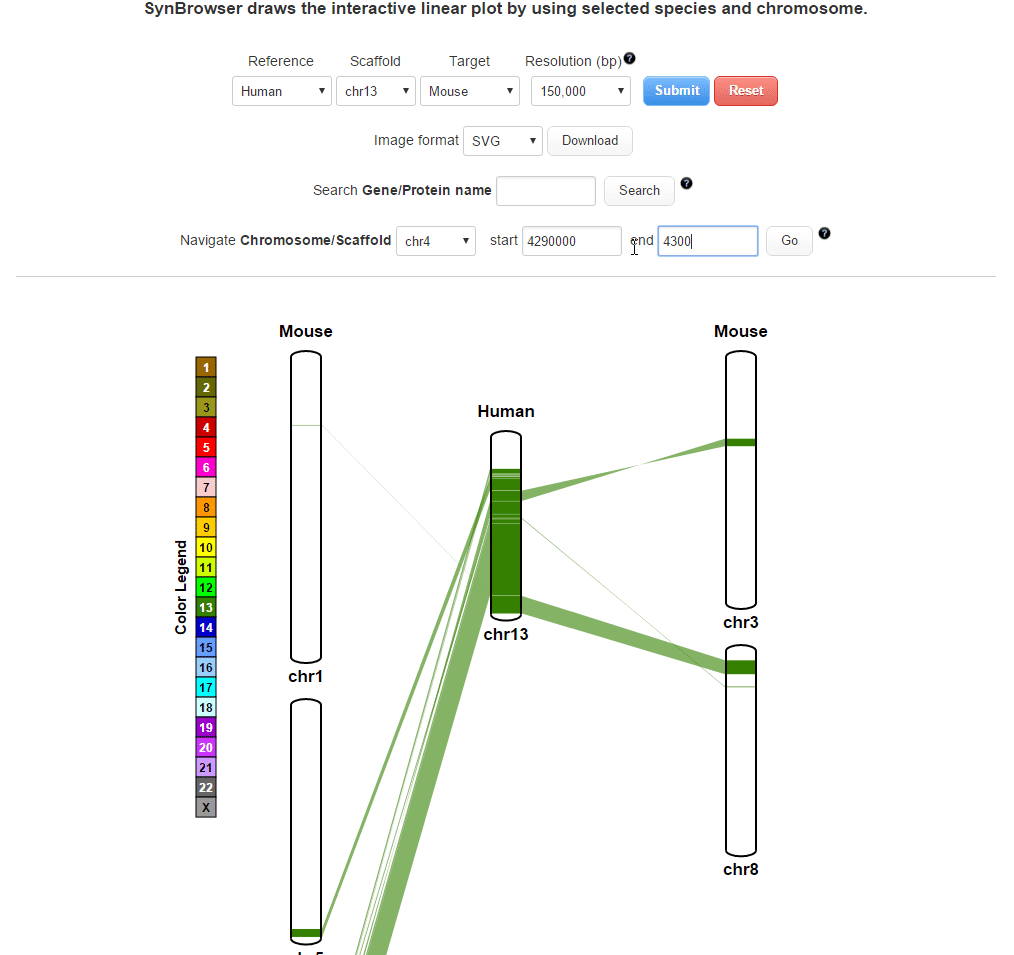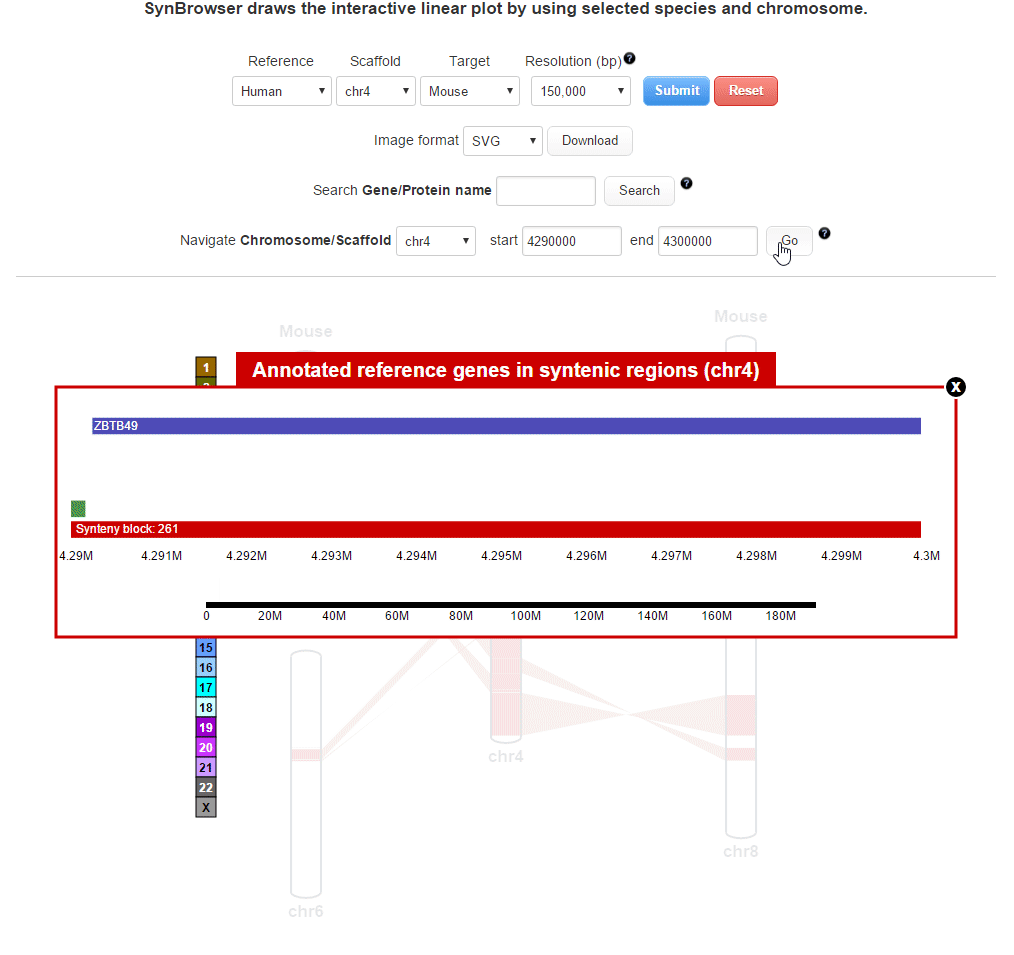
SynBrowser
SynBrowser shows synthenic relationships between two chosen species with annotated genes of a reference species.User can easily navigate the reference chromosomes by using coordinates or genes.
| 1. Drawing a plot | |
|---|---|
| (1) Selecting a reference species and a chromosome. | |
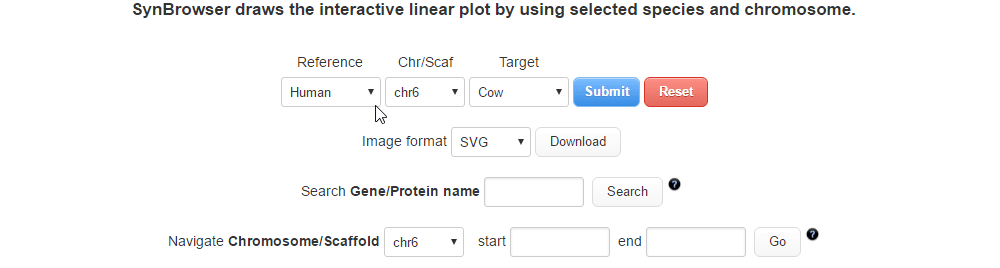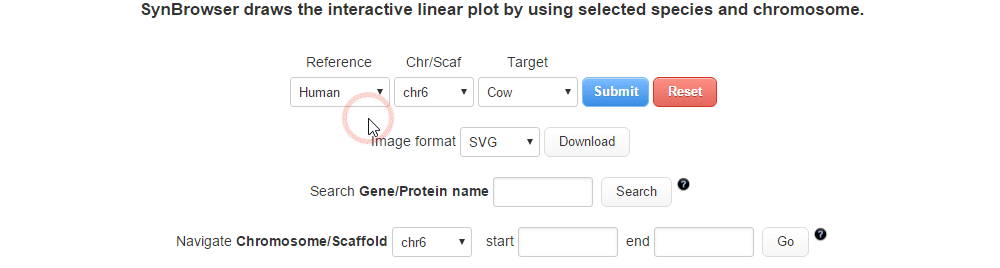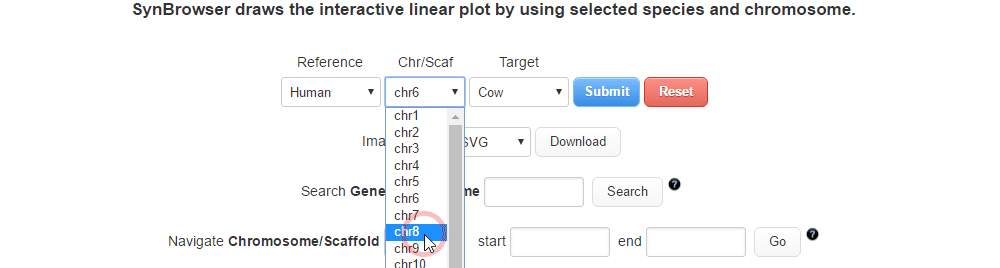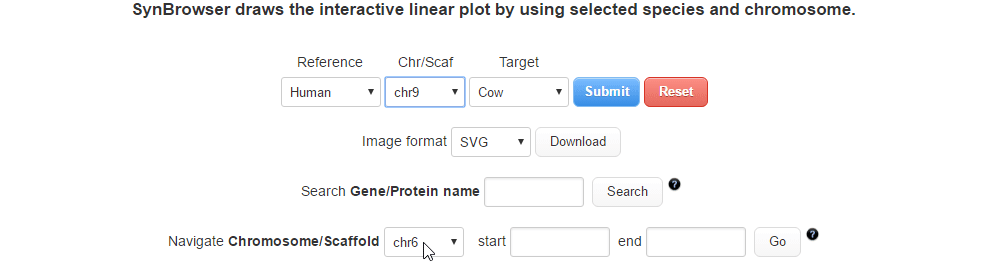 | |
| (2) Selecting a target species. | |
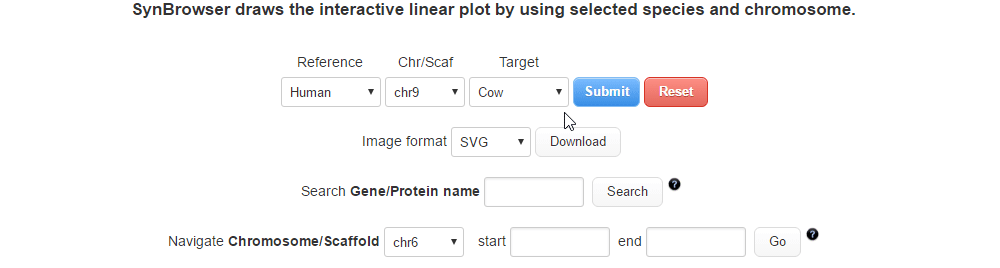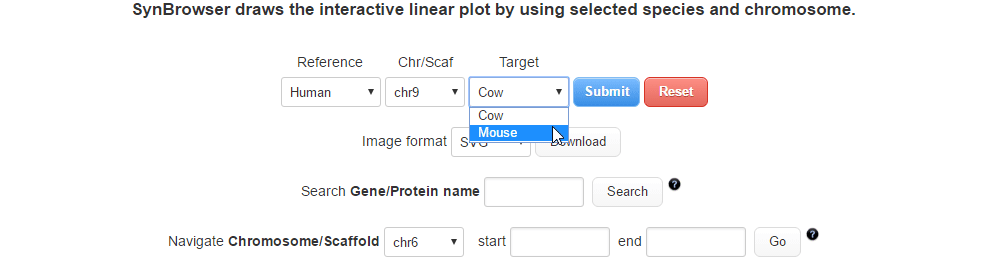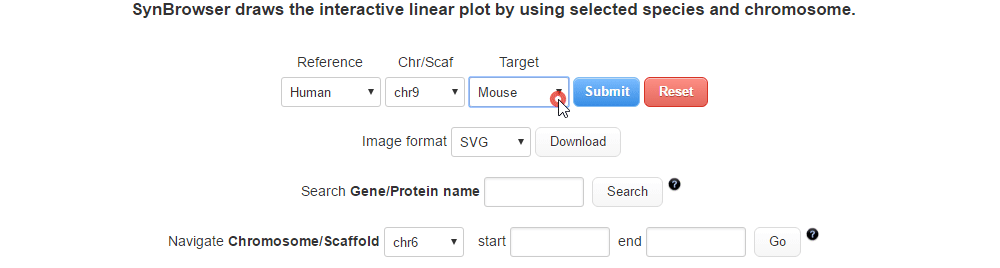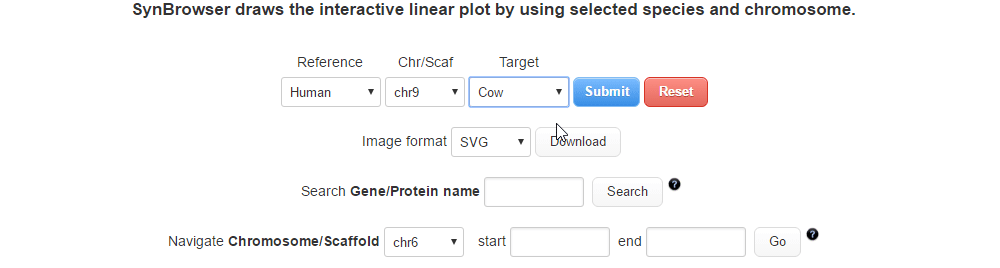 | |
| (3) Selecting a resolution of synteny blocks. (Only assemblies input) | |
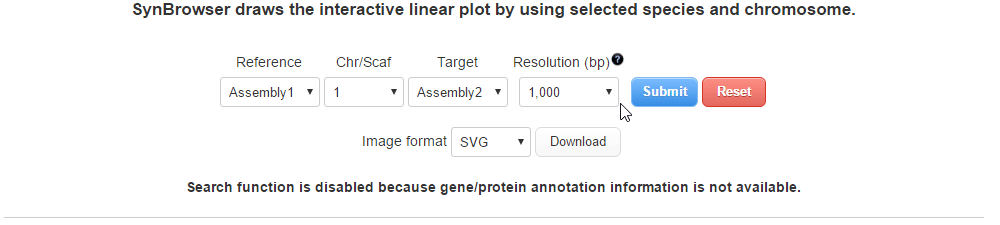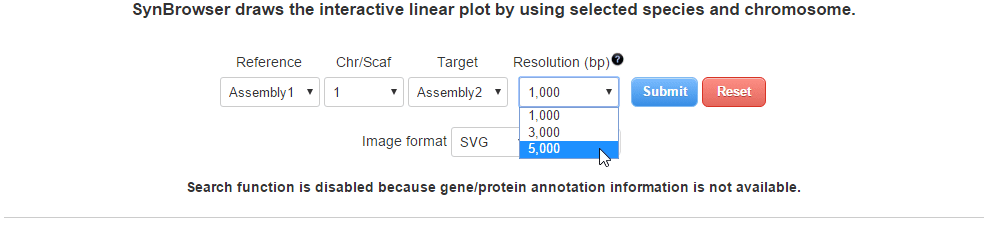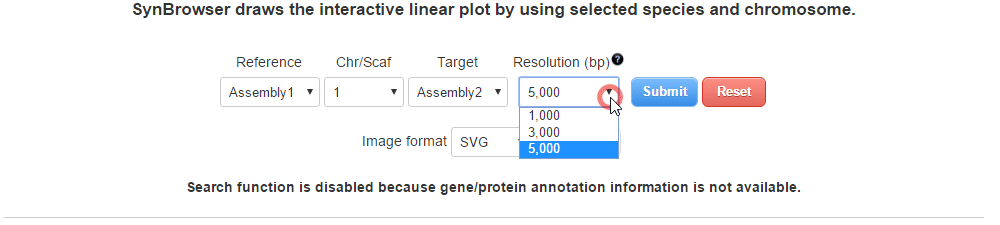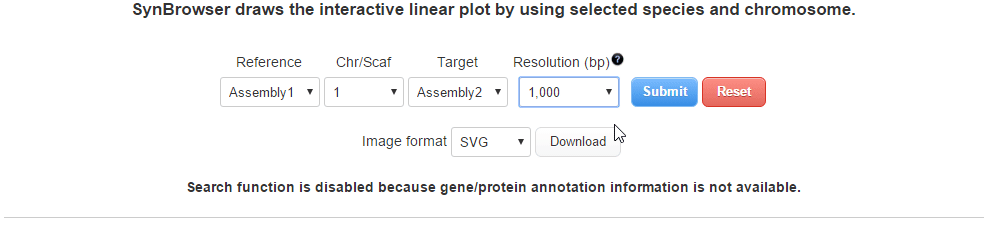 | |
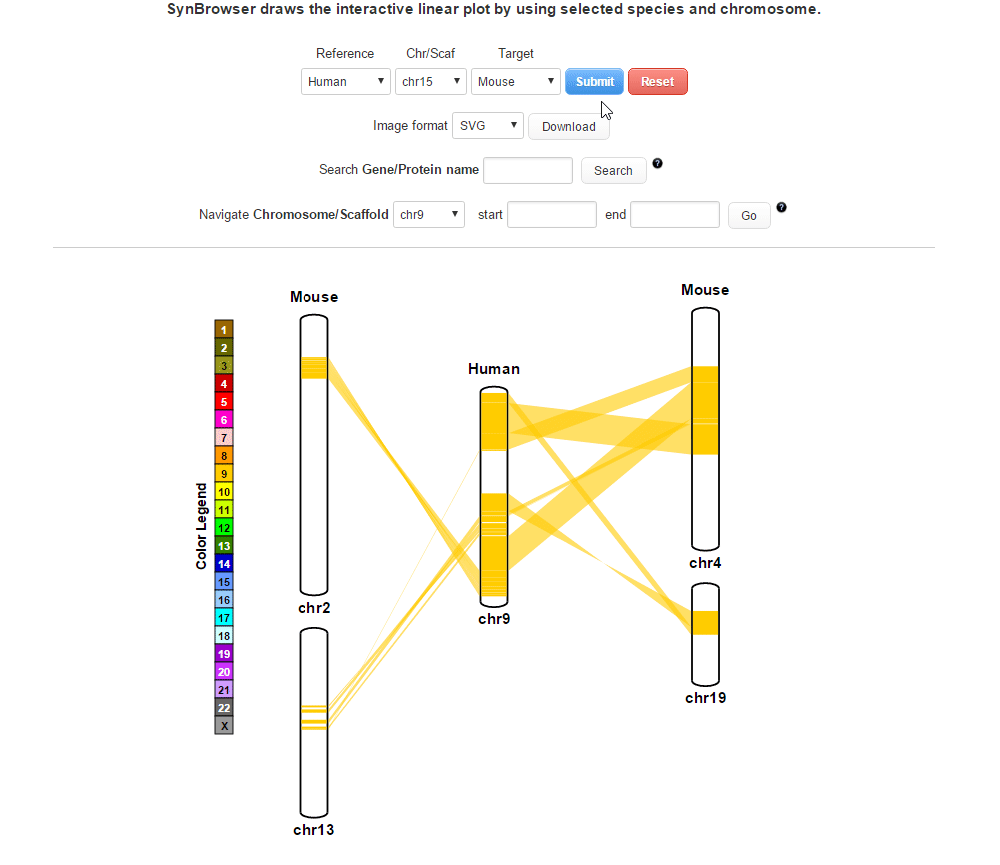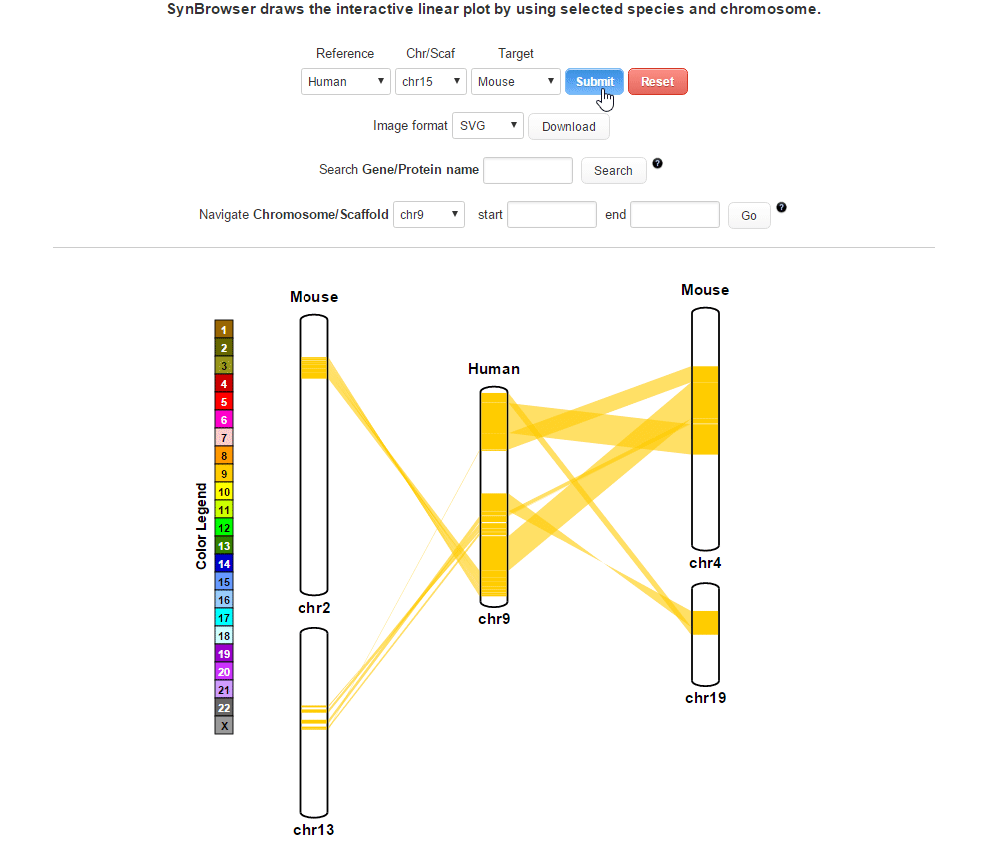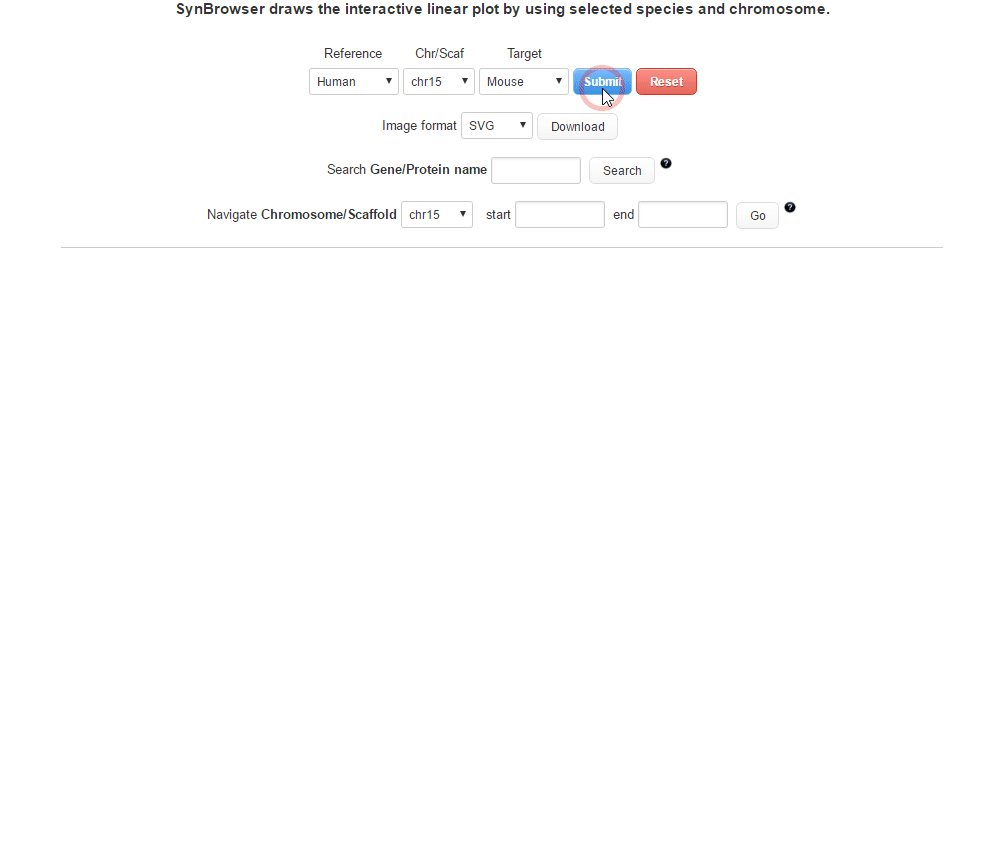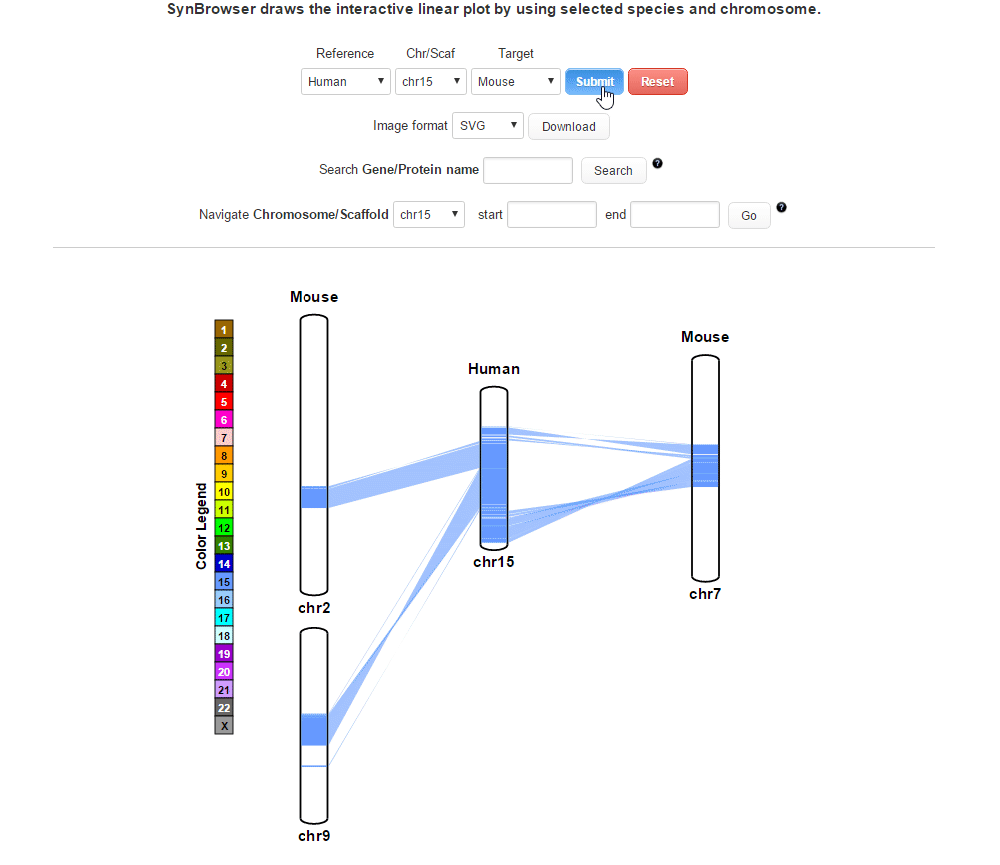
| (4) Clicking 'Submit' button to draw a plot showing pairwise synthenic information. | |
 | |
| 2. Downloading a plot | |
|---|---|
| (1) Selecting an image format. | |
 | |
| (2) Clicking on 'Download' button. | |
 | |
| 3. Browsing the details of synteny blocks | |
|---|---|
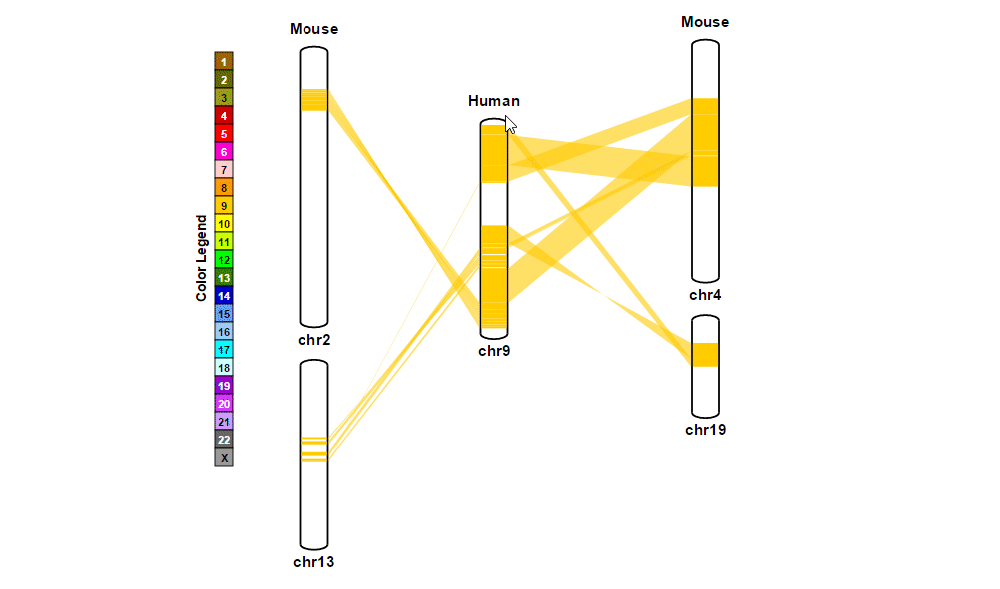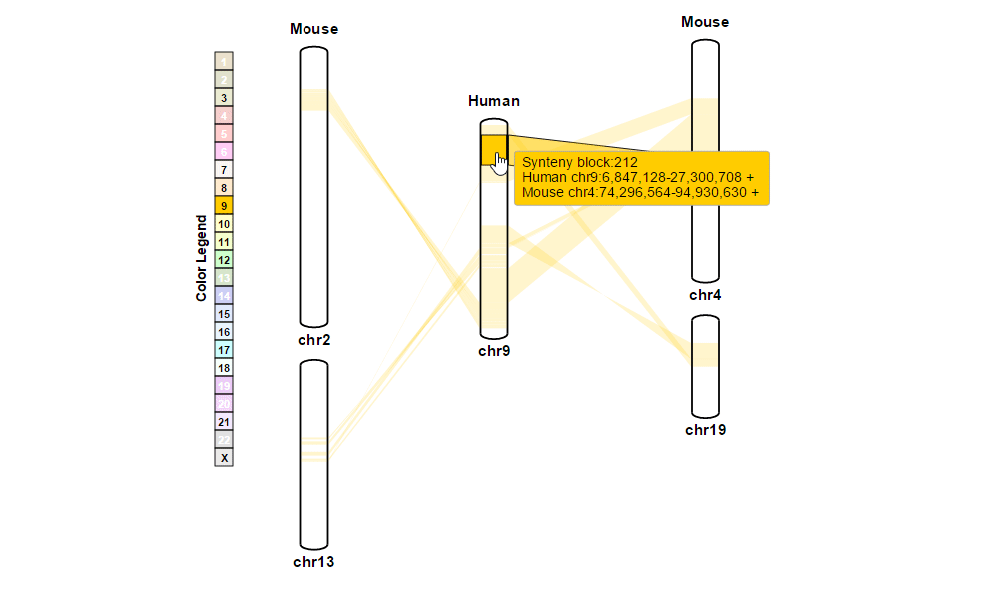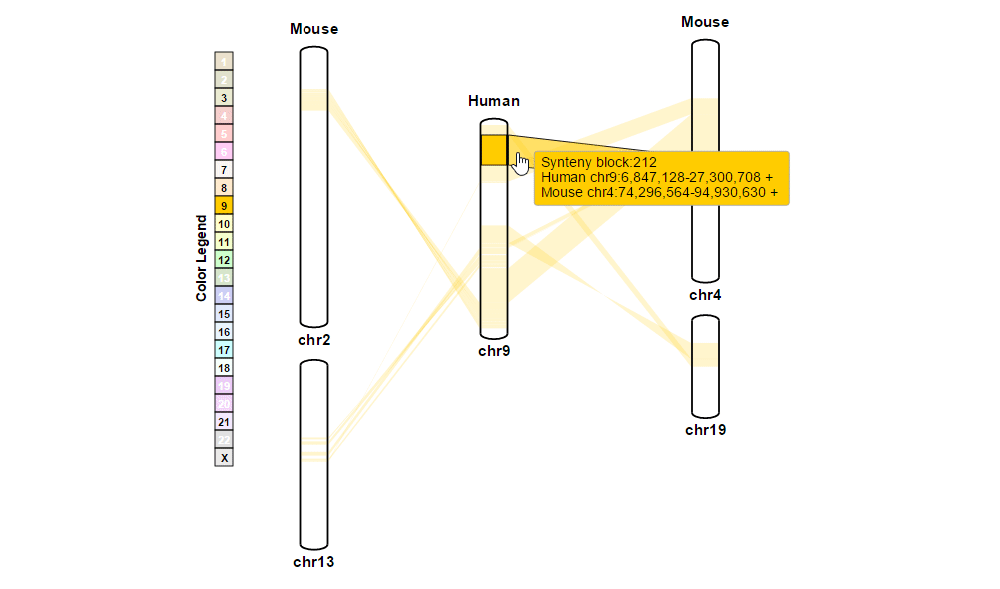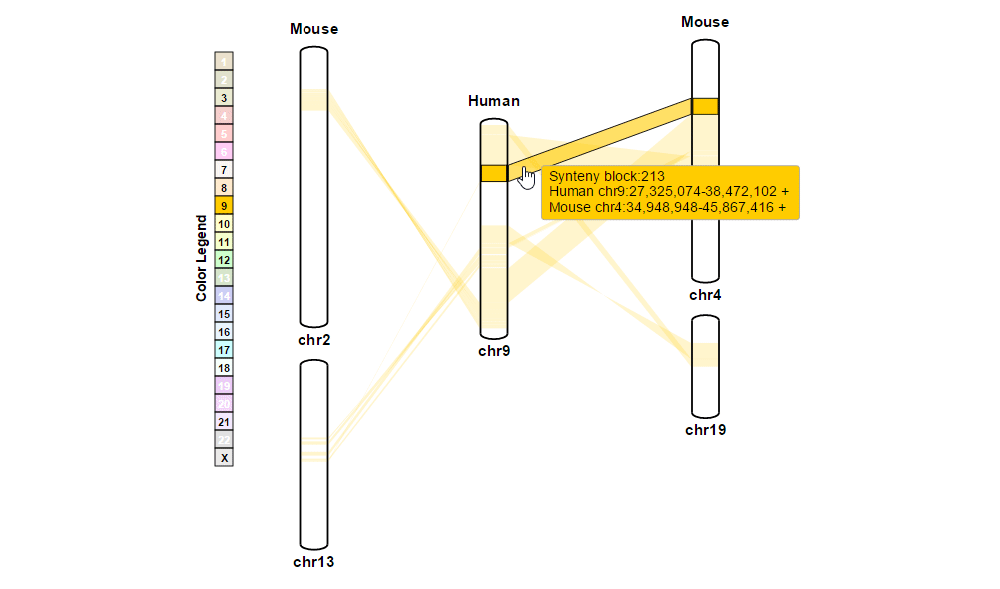
| (1) Obtaining information about synteny blocks | |
 | |
| (2) Browsing gene annotation | |
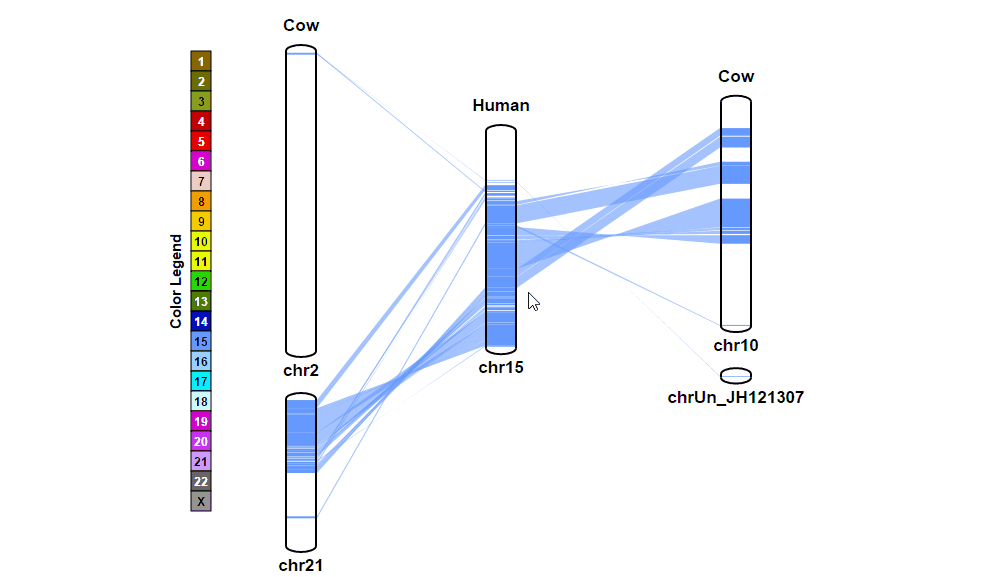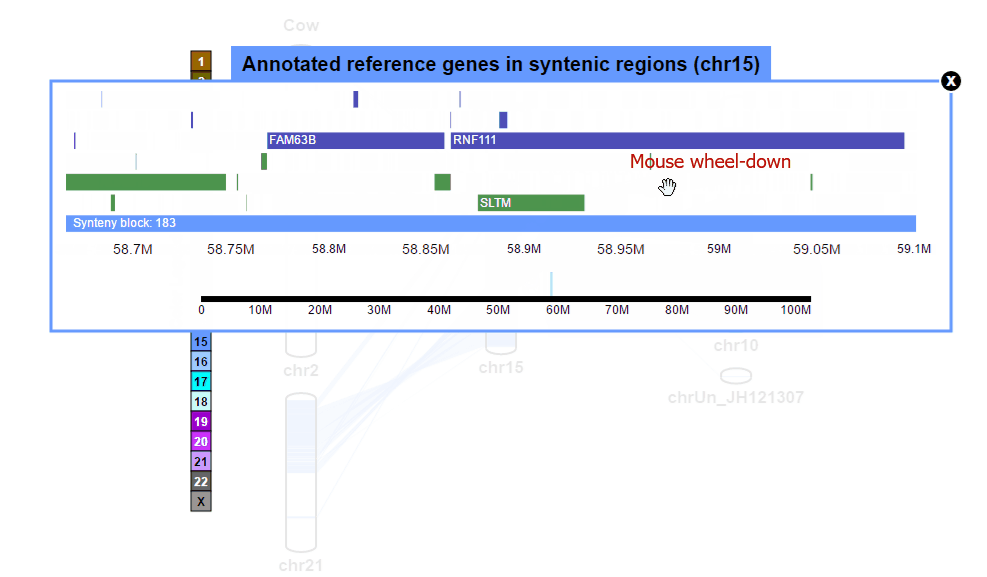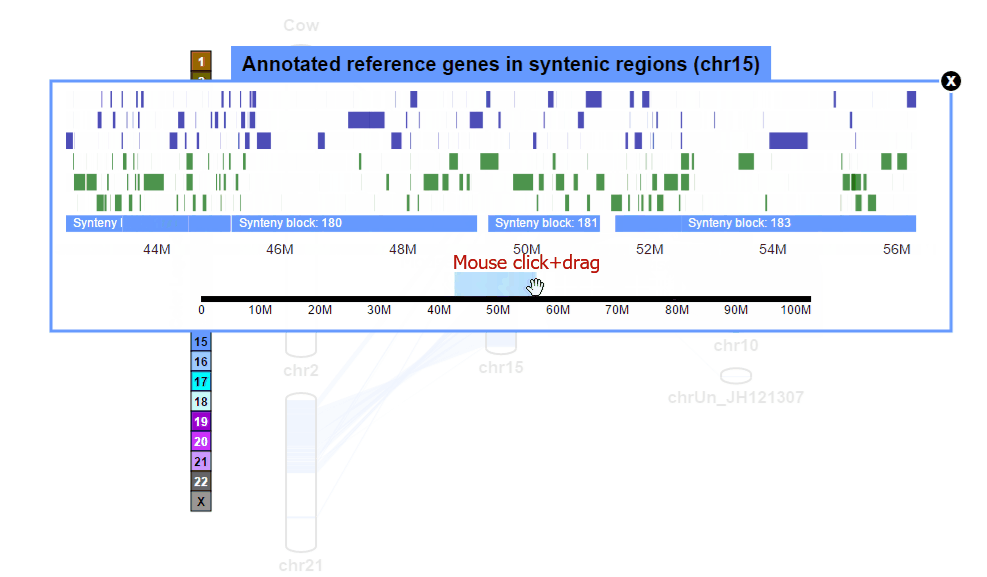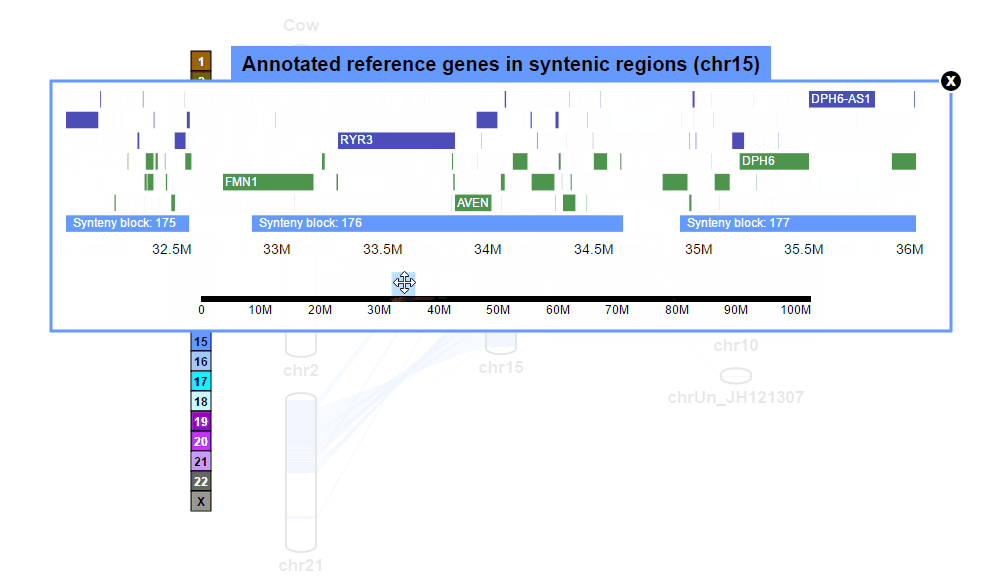 | |
| (3) Obtaining gene information | |
 | |
| 4. Searching for a specific position in synteny blocks | |
|---|---|
| (1) Selecting a reference chromosome | |
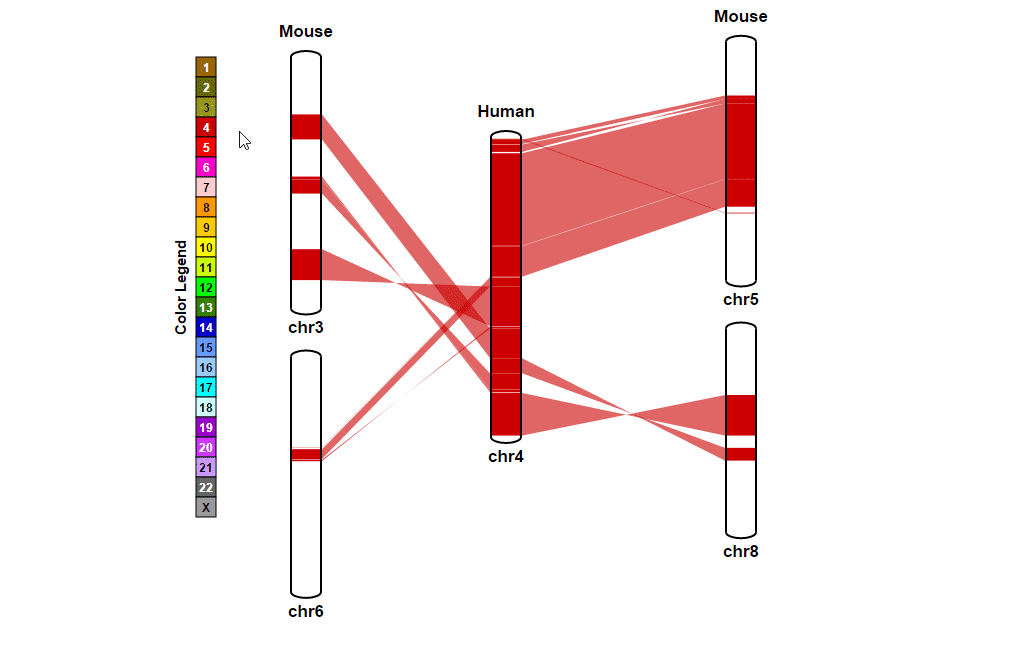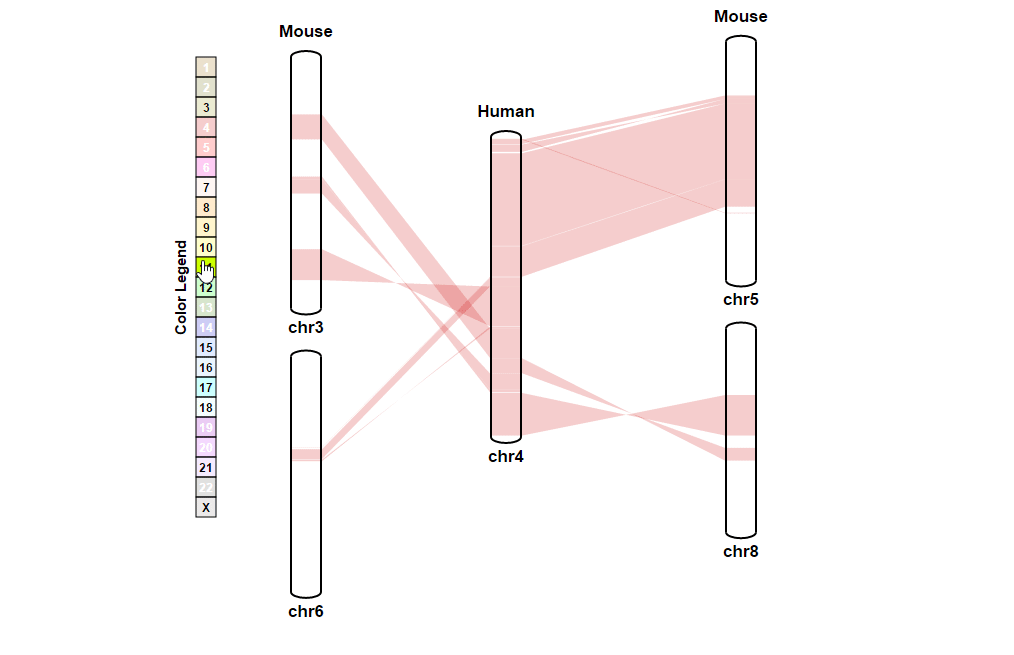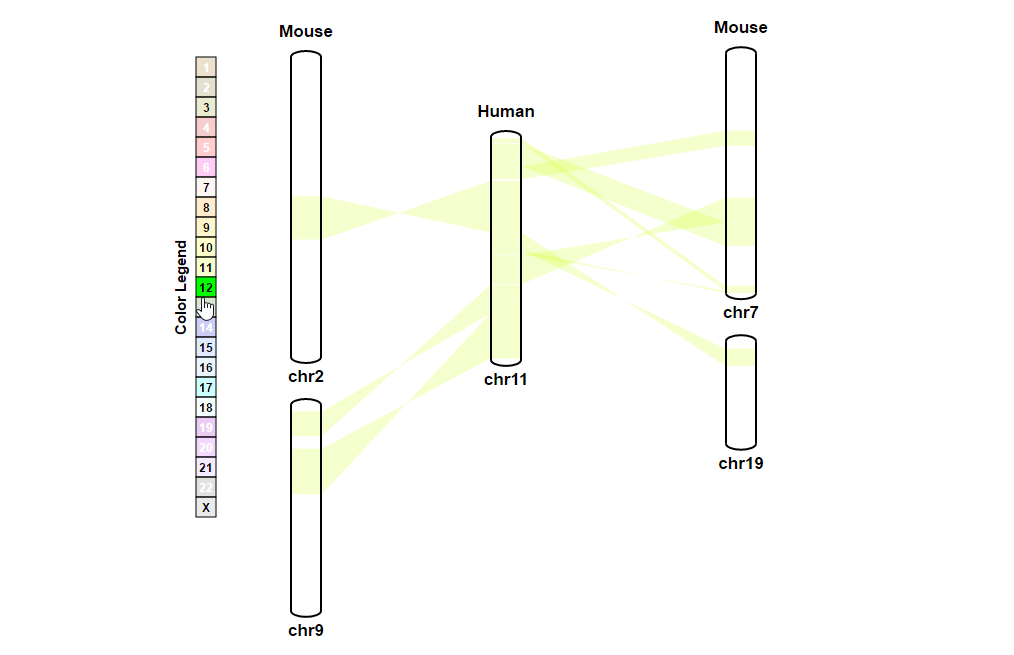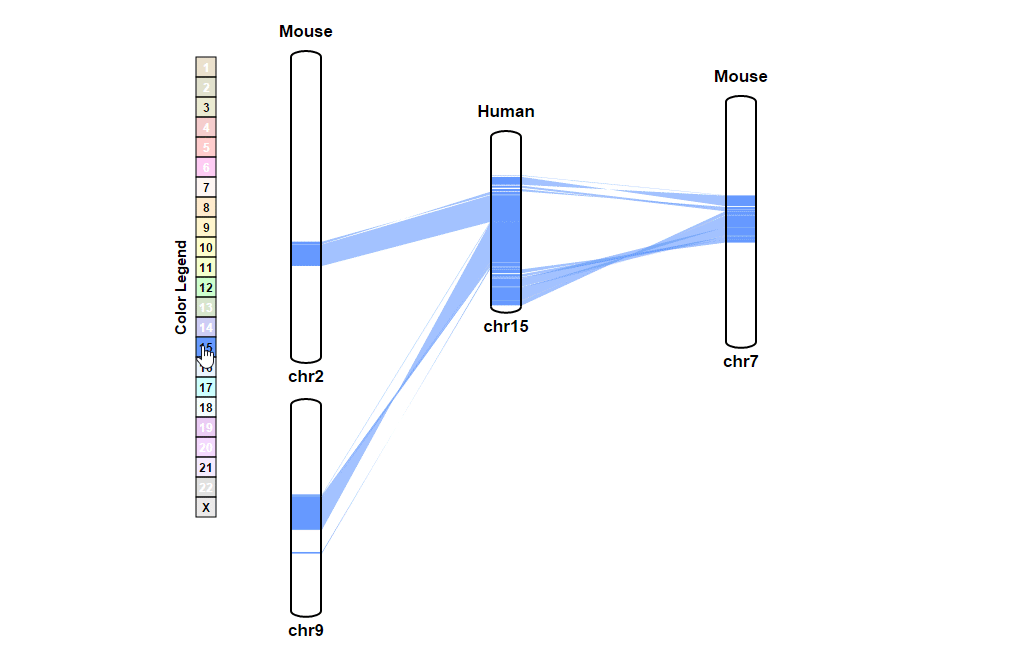 | |
| (2) Searching by using a query gene | |
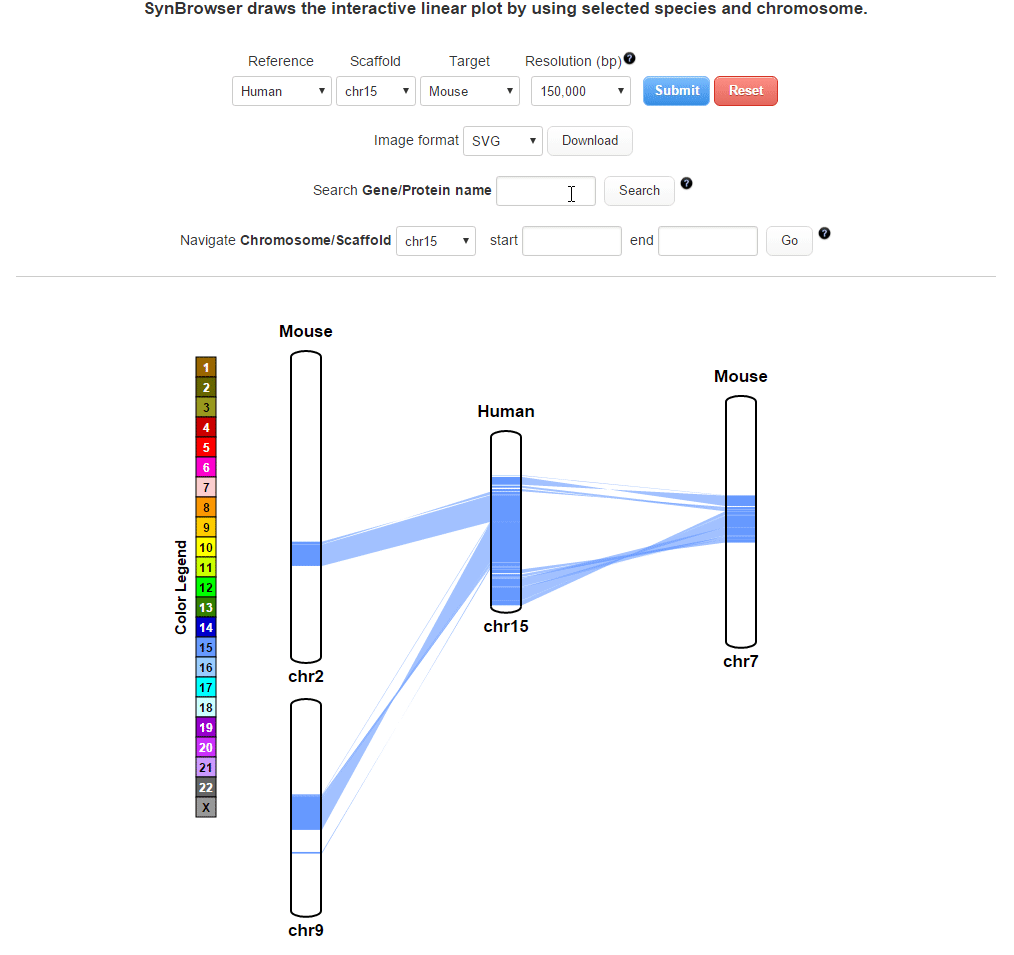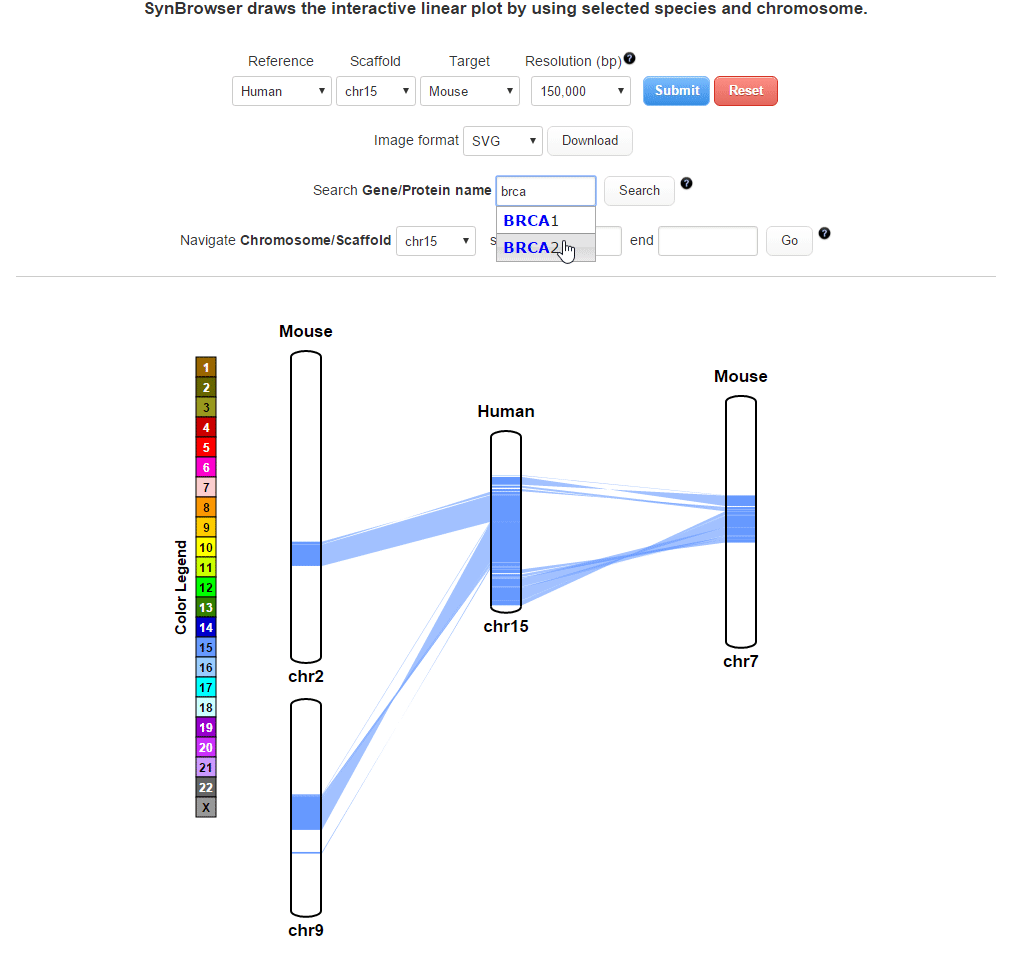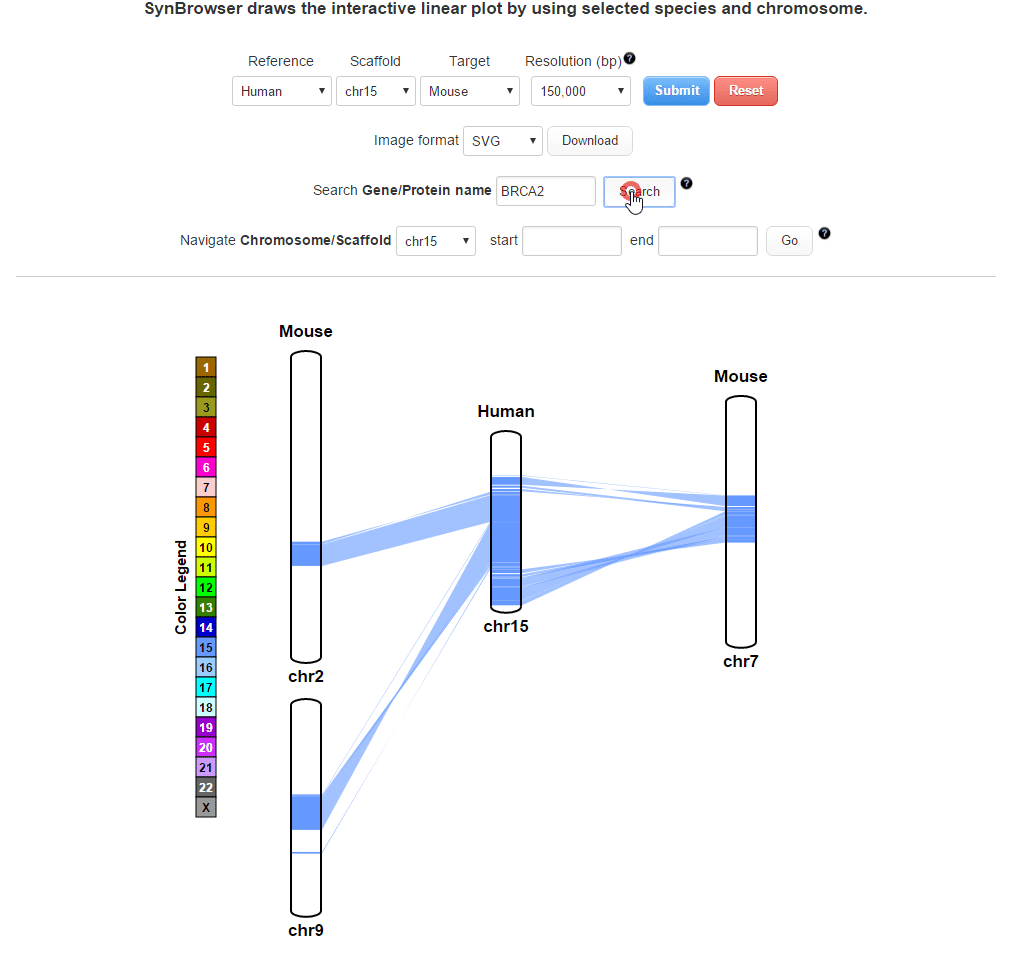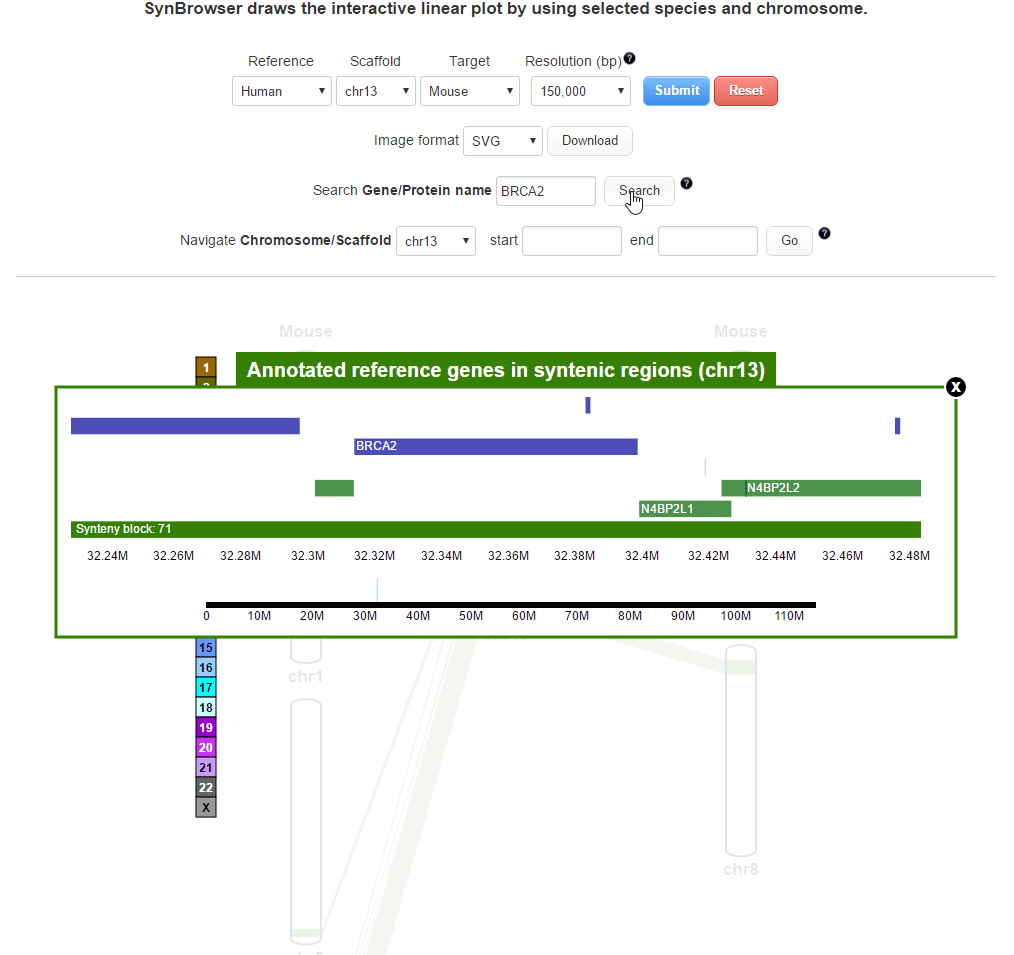 | |
| (3) Searching by using a coordinate of synteny blocks | |
 | |